Bioprospecting of Endophytic Fungi as Promising Anti-MRSA Agents
Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is a serious threat to the global healthcare system. Ineffective and resistance to antibiotic treatments have increased morbidity and mortality rates worldwide. New and effective antibiotics are needed to combat against bacterial resistance. Endophytic fungi are crucial reservoirs of novel bioactive metabolites. In particular, the secondary metabolites show promising therapeutic potential, notably, antibacterial. This review discussed the emerging potential of endophytic fungi as anti-MRSA agents. The ecological sources of endophytic fungi were discussed with the synthesis of bioactive metabolites. The mode of antibacterial actions was elucidated to give a better understanding of the mechanisms involved. This review may serve as an important reference for future discovery and developments of anti-MRSA agents from endophytic fungi.
Keywords:
Antibiotic, bioactive metabolites, endophytic fungi, methicillin-resistant, MRSA, Staphylococcus aureusIntroduction
Methicillin-resistant Staphylococcus aureus (MRSA) is an antibiotic-resistant pathogenic bacteria in the global healthcare system and community.1 According to the Global Antimicrobial Resistance Surveillance System report, bloodstream infections carried a 24.9% median resistance rate for MRSA.2 In Malaysia, 14.9% of MRSA isolates were identified from the total S. aureus clinical samples, based on the report of National Antibiotic Resistance Surveillance in 2020.3 First recorded in the 1960s, MRSA showed a high incidence rate in Asian countries, with more than 70% detected from clinical S. aureus isolates.4,5 MRSA encompasses minor skin irritation to chronic infections, such as bloodstream contamination and pneumonia.4,6 The rapid evolution of MRSA in response to antibiotics has posed therapeutic difficulties in the healthcare system.7 Besides, the overuse of antibiotics in infection control has exacerbated the development of antibiotic resistance, causing high mortality and morbidity worldwide.8 Thus, there is an urgent need to search for effective anti-MRSA agents.8 The endophytic fungi appear to be a crucial source of bioactive metabolites for anti-MRSA.9,10
Endophytic fungi are diverse microorganisms that inhabit the host tissues symbiotically without harming the host.10 Endophytic fungi obtain nutrients and habitation from the host, while fungi produce bioactive metabolites to increase host’s tolerance against biotic and abiotic stresses.11 These metabolites are regarded as one of the primary bioactive natural products with various medicinal applications and potential.9,12 According to literature, approximately 75% of antibiotics are derived from the secondary metabolites of endophytic fungi.13 At least 499 new metabolites were discovered and recorded from 2017 to 2019.14 However, many endophytic fungi remain unexplored.15 Therefore, bioprospecting biologically active natural products from underexplored resources are vital for medicinal and therapeutic. Thus, this review highlighted the primary ecological sources and the bioactive metabolites produced by endophytic fungi (Table 1). Also, it discussed the mode of antibacterial actions of bioactive metabolites for a better understanding of the mechanisms involved.
Ecological Sources of Endophytic Fungi
Since the first isolation of endophytes from the ryegrass seed (Lolium temulentum L.) in 1898, endophytic fungi have been isolated from various sources.16,17 The following sections discuss the primary ecological sources of endophytic fungi, namely, terrestrial, marine, mangrove, desert, and freshwater ecosystems.18
Terrestrial – Endophytic fungi synthesized bioactive metabolites to improve the defense of hosts against pests or pathogens attacks.19 Terrestrial endophytic fungi occur primarily in the tropical, subtropical and temperate regions.15 Tropical rainforests contain a high diversity of endophytic fungi.20 Endophytic fungal association occurs in over 300,000 terrestrial host-plant species, such as higher plants from the families Hypericacae, Guttiferae, and Gentianaceae, and wild species of cool-season grasses.21 Endophytic fungi are polyphyletic organisms. They occur in different healthy tissues, including foliage, xylem, bark, and root of living plants above or under the ground.22 Most of the endophytes inhabit the intracellular spaces without causing harm to the host plants.23 In general, endophytic fungi from terrestrial ecosystems belong to the phylum Ascomycota. For instance, Phomopsis sp. PSU-H188, an ascomycete fungus in the family of Valsaceae, was isolated from the midrib of the tropical rubber tree Hevea brasiliensis in Thailand.24
Various parts of medicinal plants possess significant pharmacological properties. The synthesis of bioactive secondary metabolites was believed due to the long-term co-evolution between the host medicinal plants and endophytic fungi.25 Thus, many metabolites produced by endophytic fungi from terrestrial medicinal plants exhibit promising biological activities.26 Approximate 50% of new medications were developed from natural products since 1994.27 In this aspect, endophytic fungi can be cultured to produce natural products, relieving the need of depending on the host medicinal plants.22 Thus, the association between medicinal plants and endophytic fungi should be fully used to produce novel bioactive metabolites.23 For instance, endophytic Lasiodiplodia pseudotheobromae and Aspergillus micronesiensis were isolated from the medicinal herb Ocimum sanctum and the Chinese medicinal plant Phyllanthus glaucus, respectively, to produce antimicrobial metabolites.28,29
Marine – Marine organisms are intrinsically tolerant to extreme conditions such as salinity, excessive competition, acidification, limited resource, pollution, and climate change owing to their distinctive morphological, physiological characteristics, and symbiotic relationship with fungal endophytes.30,31 Symbiotic relationships between marine organisms and endophytic fungi play a vital role in ecological adaptation.32 Numerous endophytic fungi were isolated from marine organisms, of which 17% were from algae (seaweeds), 12% from Porifera (sponges), and 10% from Cnidaria (corals).33,34 Marine algae are the largest source of marine fungal endophytes in producing bioactive metabolites for various medicinal purposes.31 Extracts from various marine genera, such as Alternaria, Fusarium, and Penicillium, exhibit a broad spectrum of antimicrobial activities.35 For instances, Aspergillus fumigatus AF3-093A was derived from brown alga Fucus vesiculosus, while Pestalotiopsis sp. was isolated from coral Sarcophyton sp.34 Both fungal endophytes produced metabolites with potent antibacterial activity against MRSA. Among the 700 metabolites isolated from 105 marine endophytic fungi, 40% of metabolites showed antimicrobial activity, and 15% of new antimicrobial structures were identified.33
Mangrove – Mangrove forest is established in the estuary, delta, lagoon, and backwater areas in tropical and subtropical regions.36 The endophytic fungi are essential for mangrove plants to adapt and withstand harsh environmental conditions, such as changing tides, temperature and pH changes, high salt concentration, muddy, acidic, and anaerobic soils.37,38 Mangrove endophytic fungi are getting attention for their bioactive metabolites.39 Approximately 200 metabolites were isolated from mangrove plants, and the leaves possess the most diverse fungal endophyte community.36 For instance, 778 fungal isolates were reported from the leaves of Rhizophora mucronata grown at Matang Mangrove Forest Reserve of Malaysia.40 Genera of endophytic fungi commonly found in mangrove ecosystems are Alternaria, Aspergillus, Fusarium, Monochaetia, Pestalotia, Pestalotiopsis, Pithomyces, and Taxomyces.41 The majority of mangrove endophytic fungi possess chemical metabolites such as phenolics, steroids, tannins, triterpenes, alkaloids, saponins, and flavonoids with crucial impacts on therapeutic potency.36,42 For instance, polyketides (PKs) and phenolics isolated from endophytic Phoma sp. (host: Kandelia candel) and Nigrospora sp. MA75 (host: Pongamia pinnata) showed promising antibacterial activities against MRSA.43,44
Desert – Desert plants can adapt to harsh environmental situations, such as infrequent precipitation, high temperature, deficiency of water and nutrient, and intense solar radiation.45,46 They have specialized morphology, physiology, metabolic pathway, fungal symbiont and gene regulation to allow them to survive. Desert plants do not function as autonomous organisms. They have unique niches for diverse communities of associated fungi as their resistance strategy.45 Desert endophytic fungi exhibit lower diversity but higher tissue colonization rates when compared to tropical endophytic fungi.47 A meta-analysis reported that 88.73% of culturable fungal endophyte isolated from desert plants belonged to the phylum Ascomycetes. The remainders consisted of the sterile forms of the phylum Zygomycota and Basidiomycota.47 The genera Alternaria, Phoma, and Aureobasidium have the highest number of fungal isolates in desert plants.47 For instance, Cytospora chrysosperma HYQZ-931 was isolated from the desert plant Hipophae rhamnoides.48,49
Freshwater – Rivers, lakes, and streams are some of the freshwater ecosystems with crucial ecological roles and phylogenetically diverse fungal species.50 Aquatic plants, macrophytes, hydrophytes, and many riparian trees species are well-adapted to freshwater environments. They possess unique morphological and physiological features and fungal symbionts.51 Unfortunately, freshwater endophytic fungi and their mutualistic relationships with other aquatic plants are poorly known.22 Aquatic fungi often possess specialized structures, such as thin cuticles, stomata, aerenchymatous tissues, and roots. Likewise, aquatic plant contain tissues that are submerged and exposed to air.52 Submerged macrophytes genera, such as Apium, Potamogeton, and Ranunculus, usually host fungal endophytes.22 Meanwhile, arbuscular mycorrhizal fungi and dark septate endophytes are endophytic fungi often found in the roots of aquatic plants.22
Anti-MRSA Metabolites from Endophytic Fungi
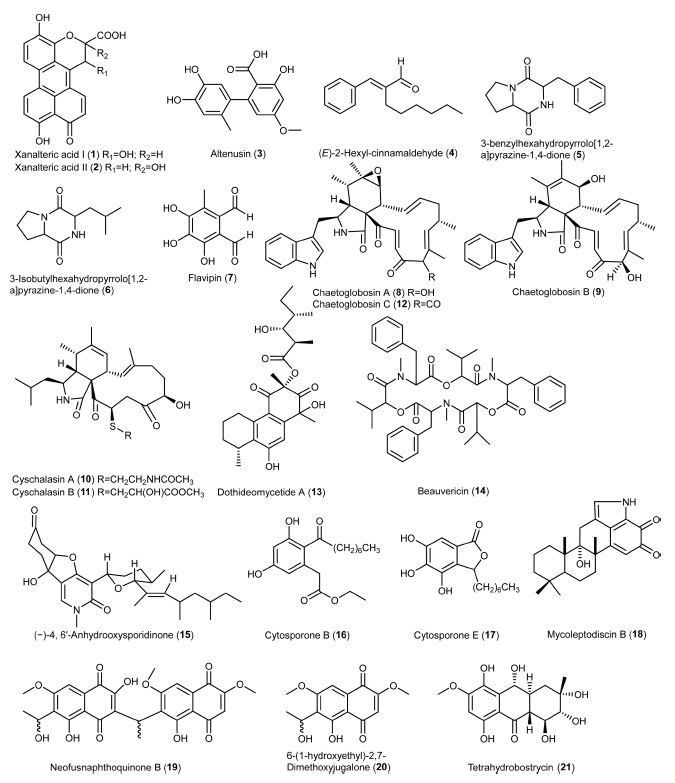
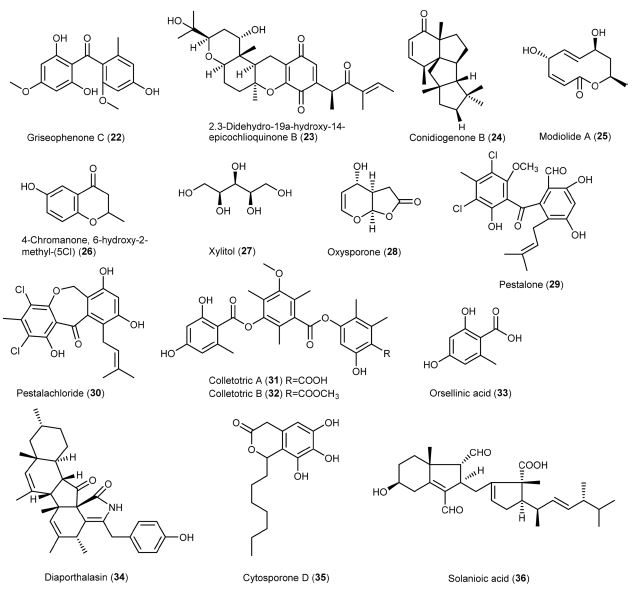
Numerous metabolites produced by endophytic fungi possess biological activities in response to environmental stress or for plant defense mechanisms.53,54 The metabolites are categorised into PKs, alkaloids, phenolics, terpenoids, etc.53,55 PKs are the largest group, followed by alkaloids and other classes. The following section discusses the various anti-MRSA metabolites isolated from endophytic fungi (Fig. 1-2).
PKs – PKs are the most abundant class of metabolites produced by endophytic fungi. They have multiple functional groups of β-hydroxy ketone or β-hydroxy aldehyde.56 PKs are synthesized through acetyl-CoA by polyketide synthases (PKS). In general, the biochemical pathway starts with incorporating acetyl-CoA units and is accompanied by a series of reactions to form different metabolites.57,58 In this context, PKS act as enzymes to catalyze the reactions of assembling PK carbon skeleton. Metabolites are modified enzymatically along the pathway via reduction and cyclization. Most PKs maintain the oxygen biosynthesis to form highly reactive poly-β-keto intermediates, and are crystalized into six-membered aromatic or 2-pyrone rings. There are also glycosylated PKs consisting of the acyl substituents on the sugar unit structure. A wide range of chemically diverse PKs may be produced, depending on the pathways that the PKS goes through. Occasionally, hybrid chemical structures may be obtained by incorporating non-ribosomal peptides into the PKs backbone.53,56,59
Xanalteric acids I and II (1-2) were obtained from endophytic Alternaria sp. isolated from S. alba as new anti-MSRA natural products, with a minimum inhibitory concentration (MIC) value of 125 and 250 μg/mL, respectively.43 Cyschalasins are metabolites consisting of tetracyclic rings. Wu et al. (2019) isolated cyschalasins A and B (10-11) from the endophytic Aspergillus micronesiensis. Cyschalasins A and B, with a significant MIC value of 17.5 and 10.6 μg/mL, respectively, showed antibacterial activity against MRSA.28 In general, cyschalasins are formed by fusing an aspochalasin and a modified cysteine residue linked to C-20 with a sulfur atom. This chemical structure is uncommon, and it is rare for naturally occurring cyschalasins to contain sulfur.60 Cyschalasins can bind to actin filaments and exhibit various biological actions.55 Other cytochalasin derivatives, such as chaetoglobosins, also exhibited antimicrobial activity. Flewelling et al. (2015) found that chaetoglobosins A (8) and B (9) showed anti-MRSA activity, with a MIC value of 95 and 378 μM, respectively. It is worth to note that these metabolites exhibited median lethal concentration (LD50) of 30 and 85 μM, respectively.79 Dissanayake et al. (2016) isolated chaetoglobosins A (8) and C (12) from the endophytic Chaetomium globosum. Chaetoglobosin A displayed better anti-MRSA activity (MIC: 32 μg/mL) than those previously reported. By contrast, chaetoglobosin C showed weaker anti-MRSA activity (MIC: >64 μg/mL) than chaetoglobosin A.50 Diaporthalasin (34), a metabolite of the cytochalasin PKs displayed a significant MIC of 4 μg/mL against MRSA.24 A tricyclic PK, dothideomycetide A (13), consisting of a linear 6,6,6-ring system, was isolated from the endophytic Dothideomycete sp. It was active to MRSA ATCC 33591 strain with a MIC value of 256 μg/mL.29 Cytosporones of the PK family have attracted the attention of researchers due to their prominent biological potential.61 The representative metabolites, cytosporones B (16) and E (17), were isolated from the endophytic Leucostoma persoonii residing in the mangrove plant Rhizophora mangle. Cytosporone B showed a 4.2-fold reduction in bacterial viability at a MIC of 78 μM, while the complete killing of bacteria occurred at 2 × MIC. More importantly, a 2-fold reduction in biofilm formation was noticed at MIC and a 168-fold reduction was observed at 2 ×MIC. As for cytosporone E, it was potent against MRSA with a MIC value of 72 μM. It gave more than 5,000-fold reduction in bacterial viability at MIC, suggesting that it could be an excellent bactericide. Besides, the minimum bactericidal concentration (MBC) value (45 μM) of cytosporone E was lower than its MIC, suggesting that it could be a potential antibiotic.62 Kongprapan et al. (2017) investigated the endophytic Phomopsis sp. PSU-H188. They obtained cytosporones B (16) and D (35) from the mycelial extract and evaluated the antibacterial activity of these two metabolites against MRSA. Both metabolites showed significant anti-MRSA activity with a MIC value of 16 and 32 μg/mL, respectively.24 Modiolide A (25), a macrolide consisting of the lactone group, was isolated from the endophytic Periconia siamensis. It showed comparable anti-MRSA activity to the standard Penicillin G (MIC: 25 μg/mL).63,64 Oxysporone (28) was first isolated from Fusarium oxysporum, an Ascomycete fungal strain. It also occurred in other fungal genera, such as Diplodia, Pestalotia, and Pestalotiopsis.65 This metabolite possesses the structure of 4H-furo(2,3-b)pyran-2(3H)-one. It was patented as an antibiotic to treat dysentery.65 Nurunnabi et al. (2017) found that oxysporone exhibited potent antimicrobial properties against various MRSA strains (ATCC 25923, SA‐1199B, RN4220, XU212, EMRSA‐15 and EMRSA‐16), with MIC values ranging from 32 to 128 μM.37 PKs colletotrics A (31) and B (32) were isolated from the mangrove endophytic fungus Phoma sp. SYSU-SK-7. These tridepsides showed strong antibacterial activity against MRSA with MIC values of 6.28 and 3.36 μg/mL, respectively.66 In the same study, a simple aromatic PK orsellinic acid (33) was obtained. It was formed through the condensation between acetyl-CoA and malonyl-CoA.67 This metabolite showed weaker antibacterial activity (8.40 μg/mL) against MRSA than colletotrics A and B.66
Alkaloids – Alkaloids are nitrogen-containing metabolites consisting of diverse chemical entities with more than 12,000 known structures.68 They are produced by various organisms such as plants, animals, bacteria, and fungi.69 Alkaloids are pharmacologically important metabolites with various properties, such as analgesic, anticancer, antibacterial, antiviral and antifungal.70 Alkaloids 3-benzylhexahydropyrrolo[1,2-a]pyrazine-1,4-dione (5) and 3-isobutylhexahydropyrrolo[1,2-a]pyrazine-1,4-dione (6) were obtained from the ethyl acetate extract of endophytic Alternaria sp. They exhibited antibacterial activity against MRSA with a MIC value of 55.55 μg/mL.71 In particular, 3-isobutylhexahydropyrrolo[1,2-a]pyrazine-1,4-dione showed anti-biofilm property that preventing bacterial adherence, and it can be used for extensive investigations against MRSA strains.72 Anhydrooxysporidinone (15) is a fungal alkaloid commonly isolated from Fusarium oxysporum, Fusarium lateritium SSF2 and Fusarium tricinctum SYPF 7082.73-75 It showed anti-MRSA activity with a MIC value of 100 μg/mL. Dissanayake and co-workers isolated an alkaloid mycoleptodiscin B (18) from the endophytic Mycoleptodiscus sp. via ethyl acetate extract of the fungal strain using column chromatography. Mycoleptodiscin B was evaluated for its action against MRSA (ATCC 33591) with a MIC value of 32 μg/mL.76
Phenolics – Phenolics are metabolites containing an aromatic ring with one or more hydroxyl (-OH) groups. Among the fungal metabolites, phenolics are responsible for several biological actions, such as antimicrobial, antioxidant, anticancer, etc.77 Altenusin (3) was isolated from the endophytic Alternaria sp. of the mangrove plant S. alba.43 This metabolite is also produced by the fungal genus Penicillium.78 Altenusin displayed potent anti-MRSA activity with a MIC value of 31.25 μg/mL.43 Flavipin (7) was isolated from the endophytic Aspergillus fumigatus.79 The metabolite consists of three phenolic and two aldehyde groups. It was firstly isolated from Aspergillus flavipes with antimicrobial action against pathogenic bacteria, fungi, and nematodes. It showed MIC and IC50 of 510 and 250 μM, respectively, against MRSA.79,80 Neofusnaphthoquinone B (19) and 6-(1-hydroxyethyl)-2,7-dimethoxyjugalone (20) were isolated as anti-MRSA metabolites from the endophytic Neofusicoccum australe. They showed significant antibacterial activity (16 μg/mL) against MRSA (ATCC 43000).81 Shang et al. (2012) isolated bioactive metabolites from the endophytic Nigrospora sp. using three different culture media. Tetrahydrobostrycin (21) and griseophenone C (22) were isolated from a culture medium containing 3.5% NaCl, while 2,3-didehydro-19a-hydroxy-14-epicochlioquinone B (23) was obtained on a medium containing 3.5% NaI. These metabolites showed exceptional anti-MRSA activity MIC values of 2, 0.5 and 8 μg/mL, respectively, compared to ampicillin (MIC: 8 μg/mL).44 Pestalone (29) was first isolated from a marine endophytic fungus, with potent activity (MIC: 37 ng/mL) against MRSA. It was regarded as a new lead in antibiotics.82 Meanwhile, pestalachloride B (30), a pestalone derivative, is structurally similar to pestalone.83 These metabolites are mostly isolated from the endophytic fungus Pestalotiopsis sp., exhibiting strong antimicrobial activity against various strains of MRSA.82,83 Wang et al. (2017) isolated pestalone and pestalachloride B from Pestalotiopsis sp. through solid-medium cultivation. These metabolites were evaluated for anti-MRSA activity against various strains with MIC values of 6.25 - 12.50 μM and 25.00 - 50.00 μM, respectively.84
Terpenoids – Terpenoids are a broad and diverse family of naturally occurring metabolites. They are derived from the 5-carbon isoprenes and categorized based on the number of isoprene units.85 Conidiogenone B (24), a tetracyclic diterpene, was isolated from a marine-derived endophytic Penicillium chrysogenum. This metabolite belonged to the class of cyclopiane with cytotoxic properties. It showed excellent anti-MRSA activity with a MIC value of 8 μg/mL.86 Solanioic acid (36) has a rare carbon structure and is rearranged from fungisterol, a steroidal precursor.87 It was isolated from Rhizoctonia solani, a fungus derived from the medicinal plant Cyperus rotundus. It exhibited significant inhibitory actions against MRSA with a MIC value of 1 μg/mL.88
Others – Other metabolites exhibiting anti-MRSA activity were aldehyde, peptide, and sugar alcohol. Beauvericin (14), a depsipeptide, was isolated from the F. oxysporum. It is a cyclic (2R)-2-hydroxy-3-methylbutanoyl-N-methyl-L-phenylalanine trimer, displaying excellent antimicrobial, insecticidal, and cytotoxic activities. It showed strong anti-MRSA action with a MIC value of 3.125 μg/mL.75,89 Sugar alcohol, also known as a polyhydric alcohol, is composed of a sugar unit containing an alcohol group.90 An example is a xylitol (27) that has a molecular formula of C5H12O5. It was isolated from the endophytic Pestalotia sp. and assessed for its anti-MRSA activity against strains ATCC 25923, SA‐1199B, XU212, MRSA340702 and EMRSA‐15, with MIC values ranging between 64 and 128 μM.37
Mode of Antibacterial Actions against MRSA
MRSA is the resistance against penicillin-like antibiotics in S. aureus. These antibiotics include penicillin, amoxicillin, oxacillin, and methicillin. The occurrence of MRSA is often associated with the abuse of antibiotics, triggering the transformation of S. aureus into MRSA. Understanding the mode of antibacterial actions against MRSA is crucial to identifying the potential antibiotics for treating MRSA.6 S. aureus is a Gram-positive bacterium that possesses an intricate structure of cell walls consisting of peptidoglycan and polysaccharide units. The cell walls of S. aureus comprise teichoic acid integrated into the cell wall polymer covalently.91 Teichoic acids are anionic polymers consisting of polyglycerol phosphate units. Primarily, they play a role in regulating cell morphology and division.92 The cell wall structure of MRSA acts as a shield, protecting it against antibacterial agents.93
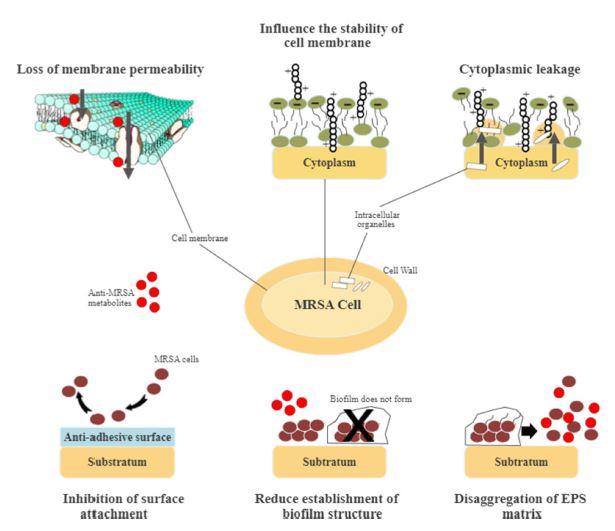
Cell wall permeability plays a vital role in bacterial cellular function, metabolic maintenance, energy transmission, osmoregulation, and immune evasion. However, the permeability of the cell wall and cell membrane could be impaired by anti-MRSA metabolites. Active metabolites form complexes by attaching to different polysaccharide units of MRSA to diffuse through the cell wall. Besides, they inhibit the production of MRSA cell walls by binding the polypeptide monomers while preventing the formation of peptide cross-links. The cell walls may then be destructed, rupturing the cell membrane.26,94-96 Surface depressions, irregular shapes, and biconcave appearances occurred on MRSA cells when treated with anti-MRSA metabolites.26,97 The permeability of the MRSA cell wall changed when examined using crystal violet uptake assay after treatment with anti-MRSA metabolites. The cell membrane of the bacterial cell wall showed a considerable decrease in crystal violet uptake due to a defective cell wall.94 Another study showed that anti-MRSA metabolites might act on cytoplasmic bacterial membrane, inducing structural malformation of the bacterial phospholipids bilayer.98,99 Thus, targeting bacterial cell wall disruption and permeability appeared as an important approach in battling against MRSA as they exerted substantial bactericidal effect.100
The integrity of bacterial cell walls and the cell membrane is vital for survival. Severe oxidation and modification of the cells could impact bacterial viability, causing a series of lipid peroxidation, protein and polysaccharide oxidation.101 A study reported the formation of cavities and tiny dents on the MRSA cell surface when treated with anti-MRSA metabolites. These observations suggested damage on the cell surface with subsequent rupture, thus producing a series of toxic products that influenced the stability of the cell membrane.96 The leakage of the cell electrolytes, intracellular organelles and ions, DNA, RNA, and proteins then occurred, impacting the metabolic pathways and enzyme activities in the MRSA cells. The osmoregulatory ability was then reduced, followed by the penetration of toxic products entering the cells, and subsequently, the loss of salt tolerance.97,101,102 Together, membrane-damaging bioactive metabolites could react to the bacterial membrane bilayer. They disrupted bacterial cell function and physical integrity, leading to the loss of membrane permeability. The bacterial membrane properties were then modified, thus, promoting the breakdown of the membrane potential.100
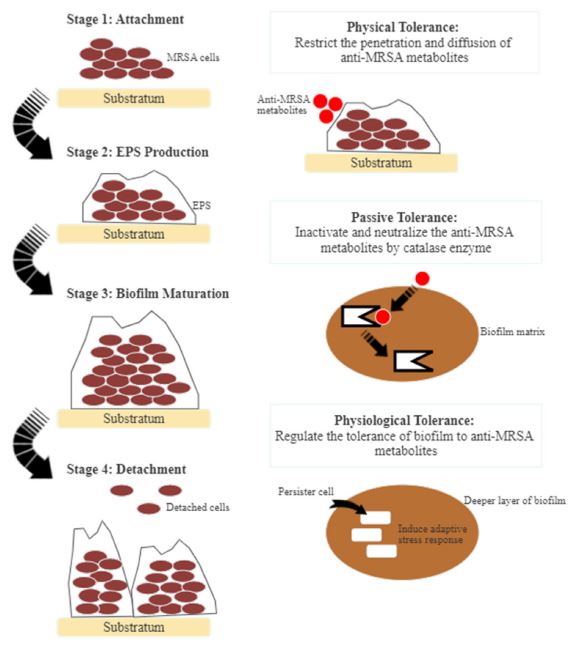
Occasionally, MRSA could respond to bioactive metabolites and counter the neutrophils through leukocytic toxins. They may use regulators to tolerate stress or cell damage exerted by antibacterial agents.103 In general, the stress response is controlled by the sigma factor B (σB), giving protection against a broad spectrum of stresses, such as acidic and alkaline conditions, heat and oxidants. This σB controls the production of virulence factors, and hence, the biofilm formation and bacterial intracellular survival.104 For instance, cell-to-cell clumping is a distinct metabolic mechanism displayed by MRSA cells to prepare for stress conditions.26 Therefore, understanding the bacterial tolerance mechanism is vital for identifying new therapeutic agents against MRSA.
MRSA strains are strong biofilm producers. They form surface-adherent communities for the cells to survive under harsh conditions, such as nutrient depletion, high pH change, and heat shock.105 Biofilms consist of bacterial cells attaching to a matrix of extracellular polymeric substance (EPS) composed of modified lipids, glycoproteins, extracellular DNA, and polysaccharides. Biofilm prevents antagonistic actions of the host cells and indirectly increases the resistance against antibacterial agents.106 Biofilm-forming is a significant virulence factor in the development of MRSA infections. MRSA pathogenicity is due to the adhesion and invasion actions that act as a barrier against antibacterial metabolites.105 The mode of actions of biofilm-forming is divisible into three different sections, i.e. physical tolerance, passive tolerance, and physiological tolerance (Fig. 3). In general, anti-MRSA metabolites eradicate biofilm by targeting different stages of formation. These stages are the attachment of bacterial cells, the development and maturation of biofilm structure, and dispersion.107 The inhibition of biofilm formation at the initial stage includes preventing the adhesion of bacterial cells and reducing the development and establishment of biofilm structure.105 Several anti-MRSA metabolites are anti-adhesive. They block adhesins, inhibit surface attachment, and eventually reduce the adhesion by modifying the surface physiochemical properties.107 Subsequently, biofilm maturation becomes irreversible with the occurrence of EPS secretion. The matured biofilm acts as a shelter and transfers nutrients to the bacterial cells, detaching and releasing these cells.108 Fig. 4 shows the general mode of actions of anti-MRSA metabolites against MRSA. Targeting the initial stage of biofilm formation is a viable anti-MRSA strategy as anti-MRSA metabolites could inhibit the biofilm more effectively than the preformed biofilm. Also, the biofilm matrix does not function as a barrier against the antibacterial agent at this juncture.107,108
Conclusion and Future Perspectives
MRSA infection has posed a serious threat to the public healthcare system by increasing morbidity and mortality worldwide. Thus, developing drugs against MRSA becomes intensive research. In this respect, endophytic fungi represent a primary component of unexplored biodiversity. They can be obtained from various ecological sources. Their bioactive metabolites show potent activity against MRSA and might serve as an alternative treatment. Future works concerning the drug discovery against MRSA may focus on extensive screening of anti-MRSA activity and bioassay-guided isolation of active metabolites from unexplored endophytic fungi. Improvement of the chemical and metabolic stability of the lead metabolites may increase their activity. In vivo experiments and clinical trials may then be conducted to examine the toxicity and the effective administration dose for developing clinically safe antibiotics.
Acknowledgments
This research was funded by the Ministry of Higher Education Malaysia for Fundamental Research Grant Scheme with Project Code: FRGS/1/2018/STG01/USM/02/3. Yee-Min Wei would like to thank Universiti Sains Malaysia for the award of Graduate Student Financial Assistance (GRA-ASSIST).
References
-
Verma, R.; Verma, S. K.; Rakesh, K. P.; Girish, Y. R.; Ashrafizadeh, M.; Sharath Kumar, K. S.; Rangappa, K. S. Eur. J. Med. Chem. 2021, 212, 113134.
[https://doi.org/10.1016/j.ejmech.2020.113134]

- World Health Organization. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report; World Health Organization: 2021.
- Institute for Medical Research. National Antibiotic Resistance Surveillance Report; Institute for Medical Research: Malaysia, 2020.
-
Khanal, A.; Sulochan, G. C.; Gaire, A.; Khanal, A.; Estrada, R.; Ghimire, R.; Panthee, S. Int. J. Infect. Dis. 2021, 103, 48-55.
[https://doi.org/10.1016/j.ijid.2020.11.152]

-
Soe, P. E.; Han, W. W.; Sagili, K. D.; Satyanarayana, S.; Shrestha, P.; Htoon, T. T.; Tin, H. H. Trop. Med. Infect. Dis. 2021, 6, 70.
[https://doi.org/10.3390/tropicalmed6020070]

-
Liu, W. T.; Chen, E. Z.; Yang, L.; Peng, C.; Wang, Q.; Xu, Z.; Chen, D. Q. Microb. Pathog. 2021, 156, 104915.
[https://doi.org/10.1016/j.micpath.2021.104915]

-
Park, B.; Liu, G. Y. Microorg. 2021, 9, 328.
[https://doi.org/10.3390/microorganisms9020328]

-
Lee, Y.; Choi, Y.; He, H.; Dodd, M. C. Environ. Sci. Technol. 2021, 55, 2541-2552.
[https://doi.org/10.1021/acs.est.0c05274]

-
Mathivanan, K.; Chandirika, J. U.; Mathimani, T.; Rajaram, R.; Annadurai, G.; Yin, H. Ecotoxicol. Environ. Saf. 2021, 208, 111567.
[https://doi.org/10.1016/j.ecoenv.2020.111567]

-
Jalil, M. T. M.; Zakaria, N. A.; Ibrahim, D. J. Pure Appl. Microbiol. 2021, 15, 1398-1408.
[https://doi.org/10.22207/JPAM.15.3.32]

-
Alsharari, S. S.; Galal, F. H.; Seufi, A. M. J. Fungi (Basel). 2022, 8, 241.
[https://doi.org/10.3390/jof8030241]

-
Gakuubi, M. M.; Munusamy, M.; Liang, Z. X.; Ng, S. B. J. Fungi (Basel). 2021, 7, 786.
[https://doi.org/10.3390/jof7100786]

-
Ma, Y.; Xu, M.; Liu, H.; Yu, T.; Guo, P.; Liu, W.; Jin, X. AMB Express 2021, 11, 111.
[https://doi.org/10.1186/s13568-021-01272-y]

- Zheng, R.; Li, S.; Zhang, X.; Zhao, C. Int. J. Mol. Sci. 2021, 22, 75.
-
Baron, N. C.; Rigobelo, E. C. Mycology 2021, 13, 39-55.
[https://doi.org/10.1080/21501203.2021.1945699]

-
Wen, J.; Okyere, S. K.; Wang, S.; Wang, J.; Xie, L.; Ran, Y.; Hu, Y. J. Fungi (Basel). 2022, 8, 205.
[https://doi.org/10.3390/jof8020205]

-
Batra, P.; Barkodia, M.; Ahlawat, U.; Sansanwal, R.; Sharma, T.; Wati, L. Int. J. Curr. Microbiol. App. Sci. 2018, 7, 1899-1911.
[https://doi.org/10.20546/ijcmas.2018.702.229]

-
Kemung, H. M.; Tan, L. T. H.; Khan, T. M.; Chan, K. G.; Pusparajah, P.; Goh, B. H.; Lee, L. H. Front. Microbiol. 2018, 9, 2221.
[https://doi.org/10.3389/fmicb.2018.02221]

-
Thomford, N. E.; Senthebane, D. A.; Rowe, A.; Munro, D.; Seele, P.; Maroyi, A.; Dzobo, K. Int. J. Mol. Sci. 2018, 19, 1578.
[https://doi.org/10.3390/ijms19061578]

-
Oita, S.; Ibáñez, A.; Lutzoni, F.; Miadlikowska, J.; Geml, J.; Lewis, L. A.; Hom, E. F. Y.; Carbone, I.; U’Ren, J. M.; Arnold, A. E. Commun. Biol. 2021, 4, 313.
[https://doi.org/10.1038/s42003-021-01826-7]

-
Khattab, A. R.; Farag, M. A. Crit. Rev. Biotechnol. 2022, 42, 403-430.
[https://doi.org/10.1080/07388551.2021.1940087]

- Sridhar, K. R. In Endophytes and Secondary Metabolite; 2019, pp 459-474.
-
Adeleke, B. S.; Babalola, O. O. J. Fungi (Basel). 2021, 7, 147.
[https://doi.org/10.3390/jof7020147]

-
Kongprapan, T.; Xu, X.; Rukachaisirikul, V.; Phongpaichit, S.; Sakayaroj, J.; Chen, J.; Shen, X. Phytochem. Lett. 2017, 22, 219-223.
[https://doi.org/10.1016/j.phytol.2017.10.002]

-
Liu, X.; Zhou, Z. Y.; Cui, J. L.; Wang, M. L.; Wang, J. H. Appl. Microbiol. Biotechnol. 2021, 105, 7095-7113.
[https://doi.org/10.1007/s00253-021-11554-x]

- Jalil, M. T. M.; Ibrahim, D. Int. J. Pharm. Pharm. Sci. 2018, 10, 50-55.
-
Calixto, J. B. An. Acad. Bras. Cienc. 2019, 91, e20190105.
[https://doi.org/10.1590/0001-3765201920190105]

-
Wu, Z.; Zhang, X.; Anbari, W. H. A.; Zhou, Q.; Zhou, P.; Zhang, M.; Zeng, F.; Chen, C.; Tong, Q.; Wang, J.; Zhu, H.; Zhang, Y. J. Nat. Prod. 2019, 82, 2653-2658.
[https://doi.org/10.1021/acs.jnatprod.9b00016]

-
Senadeera, S. P. D.; Wiyakrutta, S.; Mahidol, C.; Ruchirawat, S.; Kittakoop, P. Org. Biomol. Chem. 2012, 10, 7220-7226.
[https://doi.org/10.1039/c2ob25959a]

-
Techaoei, S.; Jirayuthcharoenkul, C.; Jarmkom, K.; Dumrongphuttidecha, T.; Khobjai, W. Saudi J. Biol. Sci. 2020, 27, 2883-2889.
[https://doi.org/10.1016/j.sjbs.2020.08.037]

-
Kamat, S.; Kumari, M.; Taritla, S.; Jayabaskaran, C. Front. Mar. Sci. 2020, 7, 31.
[https://doi.org/10.3389/fmars.2020.00031]

-
Naranjo-Ortiz, M. A.; Gabaldón, T. Biol. Rev. Camb. Philos. Soc. 2019, 94, 1443-1476.
[https://doi.org/10.1111/brv.12510]

-
El-Bondkly, E. A. M.; El-Bondkly, A. A. M.; El-Bondkly, A. A. M. Heliyon 2021, 7, e06362.
[https://doi.org/10.1016/j.heliyon.2021.e06362]

-
El-Gendy, M. M. A. A.; Mohamed, Z. K.; Hekal, N. Z.; Ali, F. M.; Yousef, A. E. M. Biotechnologia 2018, 99, 13-35.
[https://doi.org/10.5114/bta.2018.73559]

-
Sallemi, S.; Lekired, A.; Korbi, N.; Saadouli, I.; Cherif, A.; Zidi, I.; Klibi, N.; Ouzari, H.; Mosbah, A. Evid. Based Complement. Alternat. Med. 2022, 2022, 7151655.
[https://doi.org/10.1155/2022/7151655]

- Kuzhalvaymani, K.; Elizabethjacquline, L.; Subha, T. S. Int. J. Pharm. Sci. Res. 2020, 11, 4134-4143.
-
Nurunnabi, T. R.; Nahar, L.; Al-Majmaie, S.; Rahman, S. M. M.; Sohrab, M. H.; Billah, M. M.; Ismail, F. M. D.; Rahman, M. M.; Sharples, G. P.; Sarker, S. D. Phyther. Res. 2018, 32, 348-354.
[https://doi.org/10.1002/ptr.5983]

-
Kemung, H. M.; Tan, L. T.; Chan, K.; Ser, H.; Law, J. W.; Lee, L.; Goh, B. BioMed Res. Int. 2020, 2020, 6402607.
[https://doi.org/10.1155/2020/6402607]

-
Deshmukh, S. K.; Agrawal, S.; Prakash, V.; Gupta, M. K.; Reddy, M. S. South African J. Bot. 2020, 134, 237-263.
[https://doi.org/10.1016/j.sajb.2020.01.006]

-
Hamzah, T. N. T.; Lee, S. Y.; Hidayat, A.; Terhem, R.; Faridah-Hanum, I.; Mohamed, R. Front. Microbiol. 2018, 9, 1-17.
[https://doi.org/10.3389/fmicb.2018.01707]

-
Uzma, F.; Mohan, C. D.; Hashem, A.; Konappa, N. M.; Rangappa, S.; Kamath, P. V.; Singh, B. P.; Mudili, V.; Gupta, V. K.; Siddaiah, C. N.; Chowdappa, S.; Alqarawi, A. A.; Abd-Allah, E. F. Front. Pharmacol. 2018, 9, 309.
[https://doi.org/10.3389/fphar.2018.00309]

-
Kiti, H. M.; Munga, C. N.; Odalo, J. O.; Guyo, P. M.; Kibiti, C. M. West. Indian Ocean J. Mar. Sci. 2021, 20, 125-136.
[https://doi.org/10.4314/wiojms.v20i1.11]

-
Kjer, J.; Wray, V.; Edrada-Ebel, R. A.; Ebel, R.; Pretsch, A.; Lin, W.; Proksch, P. J. Nat. Prod. 2009, 72, 2053-2057.
[https://doi.org/10.1021/np900417g]

-
Shang, Z.; Li, X. M.; Li, C. S.; Wang, B. G. Chem. Biodivers. 2012, 9, 1338-1348.
[https://doi.org/10.1002/cbdv.201100216]

- Ali, A. H.; Radwan, U.; El-Zyat, S.; El-Sayed, M. Biol. Forum - An Int. J. 2018, 10, 138-145.
-
Yadav, G.; Meena, M. Biotechnol. Rep (Amst). 2021, 30, e00629.
[https://doi.org/10.1016/j.btre.2021.e00629]

-
Zhang, Q.; White, J. F. Biology (Basel). 2021, 10, 961.
[https://doi.org/10.3390/biology10100961]

-
Bashyal, B. P.; Kithsiri Wijeratne, E. M.; Faeth, S. H.; Gunatilaka, A. A. L. J. Nat. Prod. 2005, 68, 724-728.
[https://doi.org/10.1021/np058014b]

-
Mou, Q. L.; Yang, S. X.; Xiang, T.; Liu, W. W.; Yang, J.; Guo, L. P.; Wang, W. J.; Yang, X. L. Tetrahedron Lett. 2021, 87, 153207.
[https://doi.org/10.1016/j.tetlet.2021.153207]

-
Dissanayake, R. K.; Ratnaweera, P. B.; Williams, D. E.; Wijayarathne, C. D.; Wijesundera, R. L. C.; Andersen, R. J.; de Silva, E. D. Mycology 2016, 7, 1-8.
[https://doi.org/10.1080/21501203.2015.1136708]

-
Zheng, H.; Yu, Z.; Jiang, X.; Fang, L.; Qiao, M. J. Fungi (Basel). 2022, 8, 87.
[https://doi.org/10.3390/jof8010087]

-
Grossart, H. P.; Van den Wyngaert, S.; Kagami, M.; Wurzbacher, C.; Cunliffe, M.; Rojas-Jimenez, K. Nat. Rev. Microbiol. 2019, 17, 339-354.
[https://doi.org/10.1038/s41579-019-0175-8]

-
Springob, K.; Kutchan, T. M. In Plant-derived Natural Products: Introduction to the Different Classes of Natural Products; Osbourn, A. E.; Lanzorri, V. Ed; Springer, 2009, pp 3-50.
[https://doi.org/10.1007/978-0-387-85498-4_1]

-
Meena, H.; Hnamte, S.; Siddhardha, B. Adv. Endophytic Fungal Res. 2019, 2019, 145-169.
[https://doi.org/10.1007/978-3-030-03589-1_7]

-
Mousa, W. K.; Raizada, M. N. Front. Microbiol. 2013, 4, 65.
[https://doi.org/10.3389/fmicb.2013.00065]

-
Gomes, E. S.; Schuch, V.; de Macedo Lemos, E. G. Braz. J. Microbiol. 2014, 44, 1007-1034.
[https://doi.org/10.1590/S1517-83822013000400002]

-
Liu, J.; Liu, G. Methods Mol. Biol. 2018, 1848, 25-38.
[https://doi.org/10.1007/978-1-4939-8724-5_3]

-
Avalos, J.; Limón, M. C. Encyclopedia 2021, 2, 1-13.
[https://doi.org/10.3390/encyclopedia2010001]

-
Gayen, K.; Maity, S. K.; Bhowmick, T. K. Hydrocarbon Biorefinery; Elsevier: USA, 2022, pp 253-270.
[https://doi.org/10.1016/B978-0-12-823306-1.00005-4]

-
Ortega, H. E.; Torres-Mendoza, D.; Caballero E, Z.; Cubilla-Rios, L. J. Fungi (Basel). 2021, 7, 570.
[https://doi.org/10.3390/jof7070570]

-
Vitor, N.; Meza, A.; Gomes, R. S.; Rafique, J.; de Lima, D. P.; Beatriz, A. An. Acad. Bras. Cienc. 2021, 93, e20201347.
[https://doi.org/10.1590/0001-3765202120201347]

-
Beau, J.; Mahid, N.; Burda, W. N.; Harrington, L.; Shaw, L. N.; Mutka, T.; Kyle, D. E.; Barisic, B.; Van Olphen, A.; Baker, B. J. Mar. Drugs 2012, 10, 762-774.
[https://doi.org/10.3390/md10040762]

- Bhilabutra, W.; Techowisan, T.; Peberdy, J. F.; Lumyong, S. Res. J. Microbiol. 2007, 2, 749-755.
-
Karpinski, T. M. Mar. Drugs 2019, 17, 241.
[https://doi.org/10.3390/md17040241]

- Sarker, S. D.; Nahar, L.; Nurunnabi, T. R.; Rahman, S. M. M.; Sohrab, M. H.; Billah, M. M.; Ismail, F. M. D.; Sharples, G. P. Trends Phytochem. Res. 2017, 1, 55-60.
-
Chen, Y.; Yang, W.; Zou, G.; Chen, S.; Pang, J.; She, Z. Fitoterapia 2019, 139, 104369.
[https://doi.org/10.1016/j.fitote.2019.104369]

-
Tao, H.; Abe, I. Curr. Opin. Biotechnol. 2021, 69, 52-59.
[https://doi.org/10.1016/j.copbio.2020.11.016]

-
Richard, T.; Temsamani, H.; Cantos-Villar, E.; Monti, J. P. Adv. Botanical Res. 2013, 67, 67-98.
[https://doi.org/10.1016/B978-0-12-397922-3.00002-2]

-
Goyal, S. Ecological Role of Alkaloids. 1st ed.; Springer-Verlag Berlin Heidelberg: USA, 2013, pp 149-171.
[https://doi.org/10.1007/978-3-642-22144-6_98]

-
Thawabteh, A.; Juma, S.; Bader, M.; Karaman, D.; Scrano, L.; Bufo, S. A.; Karaman, R. Toxins (Basel). 2019, 11, 656.
[https://doi.org/10.3390/toxins11110656]

-
Tonial, F.; Maia, B. H. L. N. S.; Gomes-Figueiredo, J. A.; Sobottka, A. M.; Bertol, C. D.; Nepel, A.; Savi, D. C.; Vicente, V. A.; Gomes, R. R.; Glienke, C. Curr. Microbiol. 2016, 72, 173-183.
[https://doi.org/10.1007/s00284-015-0929-0]

-
Singh, V. K.; Mishra, A.; Jha, B. Front. Cell. Infect. Microbiol. 2019, 9, 308.
[https://doi.org/10.3389/fcimb.2019.00308]

- Lee, D.; Shim, S.; Kang, K. Biomolecules 2021, 11, 869.
-
Sun, W. J.; Zhu, H. T.; Zhang, T. Y.; Zhang , M. Y.; Wang, D.; Yang, C. R.; Zhang, Y. X.; Zhang, Y. J.Nat. Prod. Bioprospect. 2018, 8, 391-396.
[https://doi.org/10.1007/s13659-018-0171-0]

-
Wang, Q. X.; Li, S. F.; Zhao, F.; Dai, H. Q.; Bao, L.; Ding, R.; Gao, H.; Zhang, L. X.; Wen, H. A.; Liu, H. W. Fitoterapia 2011, 82, 777-781.
[https://doi.org/10.1016/j.fitote.2011.04.002]

-
Dissanayake, R. K.; Ratnaweera, P. B.; David E. Williams, D. E.; Wijayarathne, C. D.; Wijesundera, R. L. C.; Andersen, R. J.; de Silva, E. D. J. Appl. Pharm. Sci. 2016, 6, 001–006.
[https://doi.org/10.7324/JAPS.2016.600101]

-
Saranraj, P.; Behera, S. S.; Ray, R. C. Innovations in Traditional Foods; Elsevier Inc.: USA, 2019, pp 159-191.
[https://doi.org/10.1016/B978-0-12-814887-7.00007-1]

-
Cota, B. B.; Rosa, L. H.; Caligiorne, R. B.; Rabello, A. L. T.; Almeida Alves, T. M.; Rosa, C. A.; Zani, C. L. FEMS Microbiol. Lett. 2008, 285, 177-182.
[https://doi.org/10.1111/j.1574-6968.2008.01221.x]

-
Flewelling, A. J.; Bishop, A. I.; Johnson, J. A.; Gray, C. A. Nat. Prod. Commun. 2015, 10, 1661-1662.
[https://doi.org/10.1177/1934578X1501001009]

-
Ye, Y.; Xiao, Y.; Ma, L.; Li, H.; Xie, Z.; Wang, M.; Ma, H.; Tang, H.; Liu, J. Appl. Microbiol. Biotechnol. 2013, 97, 7131-7139.
[https://doi.org/10.1007/s00253-013-5013-8]

-
Cadelis, M. M.; Geese, S.; Uy, B. B.; Mulholland, D. R.; Van De Pas, S. J.; Grey, A.; Weir, B. S.; Copp, B. R.; Wiles, S. Molecules 2021, 26, 1094.
[https://doi.org/10.3390/molecules26041094]

-
Cueto, M.; Jensen, P. R.; Kauffman, C.; Fenical, W.; Lobkovsky, E.; Clardy, J. J. Nat. Prod. 2001, 64, 1444-1446.
[https://doi.org/10.1021/np0102713]

-
Augner, D.; Krut, O.; Slavov, N.; Gerbino, D. C.; Sahl, H. G.; Benting, J.; Nising, C. F.; Hillebrand, S.; Krönke, M.; Schmalz, H. G. J. Nat. Prod. 2013, 76, 1519-1522.
[https://doi.org/10.1021/np400301d]

-
Wang, C. F.; Wang, Y.; Zhang, X. L .; Wei, M. Y.; Wang, C. Y.; Shao, C. L. Chem. Nat. Compd. 2017, 53, 1174-1176.
[https://doi.org/10.1007/s10600-017-2230-x]

-
Evidente, A.; Kornienko, A.; Lefranc, F.; Cimmino, A.; Dasari, R.; Evidente, M.; Mathieu, V.; Kiss, R. Curr. Med. Chem. 2015, 22, 3502-3522.
[https://doi.org/10.2174/0929867322666150821101047]

-
Gao, S. S.; Li, X. M.; Zhang, Y.; Li, C. S.; Wang, B. G. Chem. Biodivers. 2011, 8, 1748-1753.
[https://doi.org/10.1002/cbdv.201000378]

-
Gao, H.; Li, G.; Lou, H. X. Molecules 2018, 23, 646.
[https://doi.org/10.3390/molecules23030646]

-
Ratnaweera, P. B.; Williams, D. E.; Patrick, B. O.; De Silva, E. D.; Andersen, R. J. Org. Lett. 2015, 17, 2074-2077.
[https://doi.org/10.1021/acs.orglett.5b00596]

-
Schneider, P.; Misiek, M.; Hoffmeister, D. Mol. Pharm. 2008, 5, 234-242.
[https://doi.org/10.1021/mp7001544]

-
Marcus, J. B. Culinary Nutrition; Elsevier Inc.: USA, 2013, pp 149-187.
[https://doi.org/10.1016/B978-0-12-391882-6.00004-2]

-
Sutcliffe, I. C. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 18637-18638.
[https://doi.org/10.1073/pnas.1216817109]

-
Morais, M. R.; Tian, P.; Lawless, C.; Murtuza-Baker, S.; Hopkinson, L.; Woods, S.; Mironov, A.; Long, D. A.; Gale, D. P.; Zorn, T. M.; Kimber, S. J.; Zent, R.; Lennon, R. Elife 2022, 11, e73486.
[https://doi.org/10.7554/eLife.73486]

-
Kong, E. F.; Johnson, J. K.; Jabra-Rizk, M. A. PLoS Pathog. 2016, 12, e1005837.
[https://doi.org/10.1371/journal.ppat.1005837]

- Ahmad, S. J.; Lian, H. H.; Basri, D. F.; Zin, N. M. Int. J. Pharm. Sci. Rev. Res. 2015, 30, 11-17.
- Chawawisit, K.; Bhoopong, P.; Phupong, W.; Lertcanawanichakul, M. Int. J. Pharm. Sci. Rev. Res. 2015, 35, 114-119.
-
Adnan, S. N. A.; Ibrahim, N.; Yaacob, W. A. Germs 2017, 7, 186-192.
[https://doi.org/10.18683/germs.2017.1125]

-
Agrawal, S.; Nandeibam, J.; Sarangthem, I. PLoS One 2021, 16, e02588607.
[https://doi.org/10.1371/journal.pone.0258607]

-
Carson, C. F.; Mee, B. J.; Riley, T. V. Antimicrob. Agents Chemother. 2002, 46, 1914-1920.
[https://doi.org/10.1128/AAC.46.6.1914-1920.2002]

-
Chen, C. Z.; Cooper, S. L. Biomaterials 2002, 23, 3359-3368.
[https://doi.org/10.1016/S0142-9612(02)00036-4]

-
Cote, H.; Pichette, A.; Simard, F.; Ouellette, M. E.; Ripoll, L.; Mihoub, M.; Grimard, D.; Legault, J. Front. Microbiol. 2019, 10, 2341.
[https://doi.org/10.3389/fmicb.2019.02341]

-
Zhang, X.; Sun, X.; Wu, J.; Wu, Y.; Wang, Y.; Hu, X.; Wang, X. Front. Microbiol. 2020, 11, 621.
[https://doi.org/10.3389/fmicb.2020.00621]

-
Mbekou, M. I. K.; Dize, D.; Yimgang, V. L.; Djague, F.; Toghueo, R. M. K.; Sewald, N.; Lenta, B. N.; Boyom, F. F. Biomed Res. Int. 2021, 2021, 6697973.
[https://doi.org/10.1155/2021/6697973]

-
Cheng, A. G.; DeDent, A. C.; Schneewind, O.; Missiakas, D. Trends Microbiol. 2011, 19, 225-232.
[https://doi.org/10.1016/j.tim.2011.01.007]

-
Ranganathan, N.; Johnson, R.; Edwards, A. M. Microbiol (Rwading). 2020, 166, 1088-1094.
[https://doi.org/10.1099/mic.0.000983]

-
Jalil, M. T. M.; Ibrahim, D. Malay. J. Med. Sci. 2021, 28, 24-36.
[https://doi.org/10.21315/mjms2021.28.4.4]

-
Ch’ng, J. H.; Chong, K. K. L.; Lam, L. N.; Wong, J. J.; Kline, K. A. Nat. Rev. Microbiol. 2019, 17, 82-94.
[https://doi.org/10.1038/s41579-018-0107-z]

-
Mishra, R.; Panda, A. K.; De Mandal, S.; Shakeel, M.; Bisht, S. S.; Khan, J. Front. Microbiol. 2020, 11, 566325.
[https://doi.org/10.3389/fmicb.2020.566325]

-
Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Virulence 2018, 9, 522-554.
[https://doi.org/10.1080/21505594.2017.1313372]