
Cytotoxic Activities on Human Ovarian Cancer Cell Lines by Neolignans and Diarylnonanoids from the Seed of Myristica fragrans Houtt.
Abstract
Ovarian cancer is a primary global health concern, often diagnosed at advanced stages with limited treatment options and poor prognosis. Natural products have emerged as potential sources of safe and effective therapies. From the seeds of Myristica fragrans Houtt. (nutmeg), 24 compounds, including neolignans and diarylnonanoid derivatives, were isolated and structurally elucidated. The cytotoxic activities of these isolated metabolites against cisplatin-sensitive and resistant human ovarian cancer cell lines were evaluated. In particular, myrifragranone C (23) exhibited cytotoxicity against all test cancer cell lines A2780, TOV-112D, and SK-OV3 with IC50 values of 14.1, 16.9, and 33.4 μM, respectively. Furthermore, compound 23 induced the death of A2780 and SK-OV3 cancer cells via apoptosis. Western blotting revealed that compound 23 significantly increased the expression of cleaved caspase-3 and poly-ADP ribose polymerase and promoted apoptosis via the mitogen-activated protein kinase signaling pathway. Our findings may provide a preliminary understanding of the anti-ovarian cancer effect of the active compound myrifragranone C as a potential treatment using natural products.
Keywords:
Myristica fragrans, Nutmeg, Neolignan, Diarylnonanoid, Ovarian CancerIntroduction
Ovarian cancer is the third most prevalent malignant tumor of the reproductive system and the second leading cause of cancer mortality in women worldwide.1,2 Owing to the lack of early screening and detection methods and presence of ovaries in the deep pelvic cavity without apparent symptoms, most patients with ovarian cancer are diagnosed at an advanced stage, which results in poor prognosis of this disease.3 The 5-year survival rate following diagnosis remains <50%.1,2 Cytoreductive surgery, radiation therapy, and chemotherapy are conventional treatment modalities for ovarian cancer.4 Currently, a mixture of small-molecule medicines, platinum, and/or taxane is routinely used in chemotherapy regimens for ovarian cancer.5 Chemotherapy has serious adverse effects because these drugs cannot distinguish between cancer and healthy cells.6 Therefore, high doses of these drugs are required to reach target cancer cells, increasing the risk of side effects in healthy tissue and reducing the effectiveness of the treatment.7,8 Thus, it is important to discover a highly safe therapy with good efficacy to treat patients with ovarian cancer.
Natural products have long played a vital role in the research and development of drugs for treating various diseases due to their inherent safety and lack of side effects.9,10 Myristica fragrans Houtt., also known as the nutmeg tree, belongs to the Myristicaceae family. It is an evergreen tropical tree that emits a pleasant aroma and possesses a delightful taste.11 This plant has been extensively used as both a spice and a remedy for various uman diseases in traditional and modern medicine. Over 200 secondary metabolites, including lignans, neolignans, diphenylalkanes, terpenoids, and phenolic compounds, have been isolated and structurally elucidated from M. fragrans.12 Among them, malabaricone C, a diarylnonanoid, distinguishes itself as a compound with numerous potent biological properties. Malabaricone C exhibits strong radical scavenging activity against DPPH, with an IC50 value of 6.56 µg/ml.13,14 It also demonstrates significant AChE inhibition, with an IC50 value of 2.06 µg/ml.15 Furthermore, malabaricone C exhibits promising effects in reducing systolic blood pressure, ventricular hypertrophy, and inflammation in both the ventricles and aorta of hypertensive rats induced by DOCA-salt.16
In our previous study, the isolation of 24 secondary metabolites, including 18 neolignans (1–18) and 6 diarylnonanoid derivatives (19–24), from the seeds of M. fragrans was reported.17 Herein, the cytotoxic activity of all isolated compounds (1–24) against three human ovarian cancer cell lines, A2780, TOV-112D, and SK-OV3, was further evaluated using MTT assays. Notably, strong cytotoxic effects against all tested cancer cell lines were exhibited by myrifralignan F (14), malabaricone C (20), and myrifragranone C (23). The structural elucidation of these active compounds has also been described to clarify their properties. Furthermore, additional investigations into the effects of the most potent active compound 23 on the induction of apoptosis, apoptosis markers, and mitogen-activated protein kinase (MAPK) signaling pathways in A2780 and SK-OV3 cells were carried out.
Experimental
General experimental procedures – Optical rotations were recorded on a JASCO P-2000 polarimeter (Tokyo, Japan). NMR spectra were recorded using a Bruker Avance 500 MHz spectrometer. Preparative HPLC was performed on a Waters Alliance system (Waters, USA): YMC-Pack ODS-A column (20 × 250 mm, 5 μm) and Water 2996 PDA detector. Column chromatography (CC) was performed on silica gel (Kieselgel 60, 40–63 μm) and RP-18 gel (LiChroprep, 40–63 μm) (Merck, Germany). Thin-layer chromatography was performed on silica gel 60 F254 and RP-18 F254S glass plates (Merck).
Plant material – Dried seeds of M. fragrans were obtained from the traditional market in Daegu, in February 2017. The plant authentication was verified by Professor Byung Sun Min, Daegu Catholic University. A voucher for M. fragrans seeds (code: 21A-MF) was kept at the Lab. of Pharmacognosy, College of Pharmacy, Kyungpook National University, Korea.
Extraction and isolation – Dried seeds of M. fragrans (20.0 kg) were subjected to reflux extraction with 20 L × 3 of MeOH. The resulting solution was then concentrated in vacuo. The obtained MeOH extract (3.5 kg) was suspended in 4 L of distilled water and then partitioned with n-hexane and ethyl acetate (EA). After removing the solvent, the EA extract (397.0 g) was separated by CC using CH2Cl2‒MeOH (99:1‒10:1, v/v) as the mobile phase, yielding fractions 1A‒1G. Fractions 1C (120 g) and 1D (12 g) were combined and subjected to CC with n-hexane‒EA (10:1‒1:1, v/v), resulting in the separation of seven fractions (2A‒2G). Compound 20 (3.6 g) was obtained by recrystallizing fraction 2A (9.0 g). Fraction 2D (22.0 g) was purified by passing it through a silica gel column and eluting with n-hexane‒EA (3:1, v/v), resulting in the separation of five fractions 2D1‒2D5 and the isolation of compounds 9 (289.1 mg) and 10 (17.5 mg). From fraction 2D2, RP-18 CC using MeOH‒H2O (3:1, v/v) as the eluent was conducted, led to isolate compounds 1 (6.1 g), 3 (2.8 g), and 19 (179.8 mg). Compounds 2 (67.2 mg), 4 (2.0 mg), and 5 (2.0 mg) were isolated from fraction 2D4 (715.3 mg) by HPLC using an isocratic solvent system (MeOH‒H2O, 65:35).
Fraction 2F (21.3 g) was chromatographed using RP-18 gel CC with a mobile phase of MeOH‒H2O (1:1, v/v), resulting in the separation of nine fractions 2F1–2F9. From fraction 2F4 (823.8 mg), compounds 7 (4.7 mg) and 18 (15.6 mg) were isolated using RP-18 CC with MeOH‒H2O (2:1, v/v) as the mobile phase. Using similar conditions, compounds 11 (4.4 mg), 12 (12.7 mg), and 13 (2.0 mg) were purified from 2F8 (1.34 g). From 2F6 (2.6 g), compounds 6 (158.8 mg), 8 (258.3 mg), and 14 (2.0 mg) were isolated using RP-18 CC (acetone‒H2O, 1:1, v/v). From 2F7 (618.6 mg), compounds 16 (158.8 mg) and 17 (258.3 mg) were isolated using RP-18 CC (MeOH‒H2O, 2.5:1, v/v). And from 2F9 (1.02 g), compounds 15 (2.0 mg), 21 (5.0 mg), 22 (22.0 mg), 23 (21.9 mg), and 24 (3.5 mg) were isolated using RP-18 CC (MeOH‒H2O, 5:1, v/v).
Myrislignan (1) – colorless oil; +14.0 (c 0.02, CHCl3); 1H-NMR (500MHz, CDCl3): δH 7.01 (1H, d, J = 1.7 Hz, H-2), 6.81 (1H, d, J = 8.1 Hz, H-5), 6.75 (1H, dd, J = 1.7, 8.1 Hz, H-6), 6.56 (2H, s, H-3′, H-5′), 6.01 (1H, ddt, J = 6.7, 10.0, 16.8 Hz, H-8′), 5.12 (2H, m, H-9′), 4.83 (1H, d, J = 3.3 Hz), 4.35 (1H, qd, J = 3.4, 6.4 Hz, H-8), 3.86 (3H, s, 3-OCH3), 3.85 (6H, s, 3′-OCH3, 5′-OCH3), 3.37 (2H, d, J = 6.7 Hz, H-7′), 1.16 (3H, d, J = 6.4 Hz, H-9); 13C-NMR (125 MHz, CDCl3): δC 154.6 (C-2′, C-6′), 148.6 (C-3), 146.5 (C-4), 138.6 (C-8′), 137.6 (C-4′), 134.6 (C-1′), 133.3 (C-1), 120.0 (C-6), 116.2 (C-9′), 115.7 (C-5), 111.0 (C-2), 106.8 (C-3′, C-5′), 83.7 (C-8), 75.3 (C-7), 56.3 (3-OCH3), 56.6 (2ʹ-OCH3, 6′-OCH3), 41.3 (C-7′), 13.8 (C-9).18
erythro-(7S,8R)-∆8′-7-Dihydroxy-3,4,3′,5′-tetramethoxy-8-O-4′-neolignan (2) – colorless oil; +18.8 (c 0.02, CHCl3); 1H-NMR (500 MHz, CD3OD): δH 7.04 (1H, d, J = 1.8 Hz, H-2), 6.91 (1H, d, J = 8.3 Hz, H-5), 6.87 (1H, dd, J = 1.8, 8.3 Hz, H-6), 6.58 (2H, s, H-3′, H-5′), 6.02 (1H, ddt, J = 6.7, 10.0, 16.8 Hz, H-8′), 5.14 (2H, m, H-9′), 4.84 (1H, d, J = 3.4 Hz, H-7), 4.36 (1H, qd, J = 3.4, 6.4 Hz, H-8), 3.87 (6H, s, 2′-OCH3, 6′-OCH3), 3.85 (3H, s, 3-OCH3), 3.83 (3H, s, 4-OCH3), 3.39 (2H, d, J = 7.0 Hz, H-7′), 1.16 (3H, d, J = 6.4 Hz, H-9); 13C-NMR (125 MHz, CD3OD): δC 154.7 (C-2′, C-6′), 150.2 (C-3), 149.5 (C-4), 138.7 (C-8′), 137.6 (C-1′), 134.9 (C-1, C-4′), 120.0 (C-6), 116.2 (C-9′), 112.6 (C-5), 111.4 (C-2), 106.8 (C-3′, C-5′), 83.6 (C-8), 75.3 (C-7), 56.6 (2′-OCH3, 6′-OCH3), 56.5 (3-OCH3), 56.4 (4-OCH3), 41.4 (C-7′), 13.8 (C-9).19
(7S,8R)-Rhaphidecursinol B (3) – colorless oil; +15.9 (c 0.02, CHCl3); 1H-NMR (500 MHz, CDCl3): δH 6.72 (2H, d, s, H-2, H-6), 6.59 (2H, s, H-3′, H-5′), 6.04 (1H, ddt, J = 6.8, 10.0, 16.8 Hz, H-8′), 5.15 (2H, m, H-9′), 4.87 (1H, d, J = 3.3 Hz, H-7), 4.41 (1H, qd, J = 3.6, 6.4 Hz, H-8), 3.89 (6H, s, 3-OCH3, 5-OCH3), 3.87 (6H, s, 2′-OCH3, 6′-OCH3), 3.81(3H, s, 4-OCH3), 3.40 (2H, d, J = 6.7 Hz, H-7′), 1.20 (3H, d, J = 6.4 Hz, H-9); 13C-NMR (125 MHz, CH3OH): δC 154.6 (C-3, C-5), 154.2 (C-2′, C-6′), 138.7 (C-8′), 138.1(C-4), 138.0 (C-1), 137.6 (C-1′), 134.7 (C-4′), 116.2 (C-9′), 111.0 (C-2), 106.8 (C-3′, C-5′), 104.7 (C-2, C-6), 83.4 (C-8), 75.6 (C-7), 61.1 (4-OCH3), 56.5 (2ʹ-OCH3, 6′-OCH3), 56.3 (3-OCH3, 5-OCH3), 41.4 (C-7′), 14.0 (C-9).20
erythro-(7S,8R)-3,4-Methylenedioxy-7-hydroxy-1′-allyl-3′,5′-dimethoxy)-8-O-4′-neolignan (4) – colorless oil; +26.8 (c 0.02, CHCl3); 1H-NMR (500 MHz, CDCl3): δH 6.86 (1H, s, H-5), 6.74 (2H, s, H-2, H-6), 6.45 (2H, s, H-3′, H-5′), 5.98 (1H, ddt, J = 6.8, 10.0, 16.9 Hz, H-8′), 5.93 (2H, s, -OCH2O-), 5.12 (2H, m, H-9′), 4.33 (1H, qd, J = 2.4, 6.4 Hz, H-8), 4.07 (1H, d, J = 2.4 Hz, H-7), 3.87 (6H, s, 2′-OCH3, 6′-OCH3), 3.36 (2H, d, J = 6.7 Hz, H-7′), 1.12 (3H, d, J = 6.4 Hz, H-9); 13C-NMR (125 MHz, CDCl3): δC 153.6 (C-2′, C-6′), 147.7 (C-4), 146.5 (C-3), 137.2 (C-8′), 136.3 (C-1′), 134.2 (C-1), 133.1 (C-4′), 119.3 (C-6), 116.3 (C-9′), 108.1 (C-5), 106.9 (C-2), 105.7 (C-3′, C-5′), 101.0 (-OCH2O-), 82.4 (C-8), 73.0 (C-7), 56.3 (2′-OCH3, 6′-OCH3), 40.7 (C-7′), 12.9 (C-9).21
erythreo-(7S,8R)-1-(4′-Hydroxy-3′methoxyphenyl)-1-methoxy-2-(2″-methoxy-4″-(l‴-(E)-propenyl)phenoxy) propane (5) – colorless oil; +30.1 (c 0.02, CHCl3); 1H-NMR (500 MHz, CDCl3): δH 6.95 (1H, d, J = 8.1 Hz, H-6′), 6.90 (2H, overlapped, H-2, H-3′), 6.88 (1H, dd, J = 1.6, 8.2 Hz, H-5′), 6.76 (2H, s, H-5, H-6′), 6.35 (1H, d, J = 15.7 Hz, H-7′), 6.16 (1H, m, H-8′), 5.94 (2H, s, -OCH2O-), 4.79 (1H, d, J= 2.3 Hz, H-7), 4.31 (1H, dq, J = 2.3, 6.4 Hz, H-8), 3.90 (3H, s, 2′-OCH3), 1.88 (3H, dd, J = 1.4, 6.6 Hz, H-9′), 1.17 (3H, d, J = 6.4 Hz, H-9); 13C-NMR (125 MHz, CDCl3): δC 151.7 (C-2′), 147.7 (C-4), 146.8 (C-3), 145.8 (C-1′), 134.0 (C-1, C-4′), 130.6 (C-3′), 125.2 (C-8′), 120.3 (C-6′), 119.6 (C-6), 119.2 (C-5′), 109.5 (C-3′), 108.1 (C-5), 107.1 (C-2), 101.1 (-OCH2O-), 82.7 (C-8), 73.7 (C-7), 56.0 (2′-OCH3), 18.5 (C-9′), 13.6 (C-9).22
erythro-(7R,8S)-Δ8′-4,7-Dihydroxy-3,3′,5′-trimethoxy-8-O-4′-neolignan (6) – colorless oil; ‒39.2 (c 0.02, CHCl3); 1H-NMR (500 MHz, CDCl3): δH 6.82‒6.93 (3H, m, H-2, H-5, H-6), 6.44 (2H, s, H-3′, H-5′), 5.96 (1H, ddt, J = 6.6, 9.9, 13.6 Hz, H-8′), 5.11 (2H, m, H-9′), 4.61 (1H, d, J = 8.5 Hz, H-7), 4.02 (1H, d, J = 6.2, 8.5 Hz, H-8), 3.86 (9H, s, 3-OCH3, 2′-OCH3, 6′-OCH3), 3.35 (2H, d, J = 6.6 Hz, H-7′), 1.88 (3H, d, J = 6.2 Hz, H-9); 13C-NMR (125 MHz, CDCl3): δC 152.8 (C-2′, C-6′), 146.6 (C-3), 145.4 (C-4), 137.2 (C-8′), 136.0 (C-1′), 135.4 (C-1), 132.8 (C-4′), 120.8 (C-6), 116.3 (C-9′), 114.1 (C-5), 109.6 (C-2), 105.6 (C-3′, C-5′), 86.7 (C-8), 79.2 (C-7), 56.1 (3-OCH3), 56.0 (2′-OCH3, 6′-OCH3), 40.6 (C-7′), 17.7 (C-9).23
erythro-(7R,8S)-∆8′-4,7-Dihydroxy-3,5,3′-trimethoxy-8-O-4′-neolignan (7) – colorless oil; ‒12.7 (c 0.02, CHCl3); 1H-NMR (500 MHz, CDCl3): δH 6.97 (1H, d, J = 8.6 Hz. H-6′), 6.76 (2H, m, overlapped, H-3′, H-5′), 6.59 (2H, s, H-2, H-6), 5.98 (1H, ddt, J = 6.6, 10.0, 16.8, H-8′), 5.10 (2H, m, H-9′), 4.80 (1H, d, J = 3.1 Hz, H-7), 4.31 (1H, qd, J = 3.1, 6.4 Hz, H-8), 3.88 (9H, s, 3-OCH3, 5-OCH3, 2′-OCH3), 3.37 (2H, d, J = 6.6 Hz, H-7′), 1.17 (3H, d, J = 6.4 Hz, H-9); 13C-NMR (125 MHz, CDCl3): δC 151.6 (C-3, C-5), 147.1 (C-2′), 145.0 (C-1′), 137.5 (C-8′), 135.8 (C-1), 134.0 (C-4), 131.2 (C-4′), 121.3 (C-5′), 120.2 (C-6′), 116.1 (C-9′), 112.7 (C-3′), 103.2 (C-2, C-6), 82.7 (C-8), 73.8 (C-7), 56.5 (3-OCH3, 5-OCH3), 56.0 (2′-OCH3), 40.1 (C-7′), 13.7 (C-9).24
Maceneolignan F (8) – colorless oil; ‒19.1 (c 0.02, CHCl3); 1H-NMR (500 MHz, CDCl3): δH 6.53 (2H, s, H-2, H-6), 6.44 (2H, s, H-3′, H-5′), 5.95 (1H, ddt, J = 6.8, 10.0, 16.8, H-8′), 5.10 (2H, m, H-9′), 4.77 (1H, d, J = 2.4 Hz, H-7), 4.30 (1H, dd, J = 2.4, 6.4 Hz, H-8), 3.83 (6H, s, 2′-OCH3, 6′-OCH3), 3.81 (6H, s, 3-OCH3, 5-OCH3), 3.33 (2H, d, J = 6.8 Hz, H-7′), 1.09 (3H, d, J = 6.4 Hz, H-9); 13C-NMR (125 MHz, CDCl3): δC 153.4 (C-2′, C-6′), 146.9 (C-3, C-5), 137.0 (C-8′), 136.1 (C-4′), 133.6 (C-4), 132.9 (C-1′), 131.0 (C-1), 116.1 (C-9′), 105.4 (C-3′, C-5′), 102.7 (C-2, C-6), 82.3 (C-8), 73.0 (C-7), 56.2 (3-OCH3, 5-OCH3), 56.0 (2′-OCH3, 6′-OCH3), 40.5 (C-7′), 12.7 (C-9).25
Maceneolignan H (9) – colorless oil; ‒46.9 (c 0.02, CHCl3); 1H-NMR (500 MHz, CDCl3): δH 6.87 (1H, d, J = 1.3 Hz, H-2), 6.83 (1H, dd, J = 1.3, 8.4 Hz, H-6), 6.78 (1H, d, J = 8.4 Hz, H-5), 6.38 (2H, s, H-3′, H-5′), 5.94 (1H, ddt, J = 6.7, 10.0, 16.8 Hz, H-8′), 5.85 (1H, d, J = 3.3 Hz, H-7), 5.07 (2H, m, H-9′), 4.43 (1H, m, H-8), 3.83 (3H, s, 3-OCH3), 3.81 (3H, s, 4-OCH3), 3.75 (6H, s, 2′-OCH3, 6′-OCH3), 2.15 (3H, s, CH3COO), 1.27 (1H, d, J = 6.5 Hz, H-9); 13C-NMR (125 MHz, CDCl3): δC 170.2 (CH3COO), 153.3 (C-2′, C-6′), 148.7 (C-3), 148.5 (C-4), 137.2 (C-8′), 135.7 (C-4′), 133.7 (C-1′), 130.5 (C-1), 119.2 (C-6), 115.9 (C-9′), 110.8 (C-5), 110.2 (C-2), 105.5 (C-3′, C-5′), 80.0 (C-8), 76.6 (C-7), 55.9 (2′-OCH3, 6′-OCH3), 55.8 (3-OCH3, 4-OCH3), 40.4 (C-7′), 21.2 (CH3COO), 14.4 (C-9).25
erythro-(7R,8S)-Δ8′-7-Acetoxy-3,4-methylenedioxy-3′,5′-dimethoxy-8-O-4′-neolignan (10) – colorless oil; ‒33.6 (c 0.02, CHCl3); 1H-NMR (500 MHz, CDCl3): δH 6.82 (1H, s, H-5), 6.72 (2H, s, H-2, H-6), 6.39 (2H, s, H-3′, H-5′), 5.92 (1H, 1H, ddt, J = 6.8, 10.0, 16.9 Hz, H-8′), 5.92 (2H, s, -OCH2O-), 5.82 (2H, d, J = 3.3 Hz, H-7), 5.10 (2H, m, H-9′), 4.37 (1H, dd, J = 3.3, 6.4 Hz, H-8), 3.79 (6H, s, 2′-OCH3, 6′-OCH3), 3.33 (2H, d, J = 6.6 Hz, H-7′), 2.17 (3H, s, -OCOCH3), 1.29 (3H, d, J = 6.5 Hz, H-9); 13C-NMR (125 MHz, CDCl3): δC 170.3 (-OCOCH3), 153.5 (C-2′, C-6′), 147.7 (C-4), 147.1 (C-3), 137.4 (C-8′), 135.9 (C-1′), 133.9 (C-1), 132.2 (C-4′), 120.3 (C-6), 116.1 (C-9′), 108.1 (C-5), 107.4 (C-2), 105.7 (C-3′, C-5′), 101.1 (-OCH2O-), 80.4 (C-8), 76.7 (C-7), 56.1 (2′-OCH3, 6′-OCH3), 40.6 (C-7′), 21.3 (-OCOCH3), 14.4 (C-9).21
Myrifralignan C (11) – yellowish oil; ‒40.0 (c 0.02, CHCl3); 1H-NMR (500 MHz, CDCl3): δH 6.97 (1H, d, J = 8.1 Hz, H-6′), 6.92 (1H, d, J = 1.7 Hz, H-3′), 6.90 (1H, dd, J = 1.7, 8.1 Hz, H-5′), 6.58 (2H, s, H-2, H-6), 6.36 (1H, dd, J = 1.5, 15.7 Hz, H-7′), 6.17 (1H, dq, J = 6.6, 15.7 Hz, H-8′), 4.80 (1H, d, J = 3.1 Hz, H-7), 4.32 (1H, dq, J = 3.1, 6.4 Hz, H-8), 3.90 (3H, s, 2′-OCH3), 3.88 (6H, s, 3-OCH3, 5-OCH3), 1.88 (3H, d, J = 1.5, 6.6 Hz, H-9′), 1.18 (3H, d, J = 6.4 Hz, H-9); 13C-NMR (125 MHz, CDCl3): δC 151.7 (C-2′), 147.1 (C-3, C-5), 145.7 (C-1′), 134.0 (C-4), 133.9 (C-4′), 131.0 (C-1), 130.6 (C-7′), 125.3 (C-8′), 120.0 (C-5′), 119.2 (C-6′), 109.5 (C-3′), 103.1 (C-2, C-6), 82.8 (C-8), 73.8 (C-7), 56.5 (3-OCH3, 5-OCH3), 56.0 (2′-OCH3), 18.5 (C-9′), 13.6 (C-9).20
Surinamensin (12) – colorless oil; ‒21.1 (c 0.02, CHCl3); 1H-NMR (500 MHz, CDCl3): δH 6.97 (1H, d, J = 8.1 Hz, H-6′), 6.92 (1H, d, J = 1.8 Hz, H-3′), 6.90 (1H, dd, J = 1.8, 8.1 Hz, H-5′), 6.57 (2H, s, H-2, H-6), 6.36 (1H, dd, J = 1.5, 15.6 Hz, H-7′), 6.16 (1H, dq, J = 6.6, 15.6 Hz, H-8′), 4.81 (1H, d, J = 3.2 Hz, H-7), 4.33 (1H, dq, J = 3.2, 6.4 Hz, H-8), 3.89 (3H, s, 2′-OCH3), 3.85 (6H, s, 3-OCH3, 5-OCH3), 3.82 (3H, s, 4-OCH3), 1.88 (3H, d, J = 1.5, 6.6 Hz, H-9′), 1.19 (3H, d, J = 6.4 Hz, H-9); 13C-NMR (125 MHz, CDCl3): δC 151.3 (C-3, C-5), 151.7 (C-2′), 145.7 (C-1′), 137.2 (C-4), 135.7 (C-1), 133.9 (C-4′), 130.6 (C-7′), 125.3 (C-8′), 120.1 (C-5′), 119.1 (C-6′), 109.4 (C-3′), 103.3 (C-2, C-6), 82.6 (C-8), 73.9 (C-7), 61.0 (4-OCH3), 56.3 (3-OCH3, 5-OCH3), 55.9 (2′-OCH3), 18.5 (C-9′), 13.6 (C-9).24
Myrislignanometin E (13) – colorless oil; ‒25.1 (c 0.02, CHCl3); 1H-NMR (500 MHz, CDCl3): δH 6.96 (1H, d, J = 1.7 Hz, H-2), 6.84 (1H, d, J = 8.1 Hz, H-5), 6.67 (3H, overlapped, H-6, H-3′, H-5′), 6.58 (1H, d, J = 15.8 Hz, H-7′), 6.32 (1H, dt, J = 5.7, 15.8 Hz, H-8′), 4.79 (1H, d, J = 2.7 Hz, H-7), 4.36 (3H, overlapped, H-8, H-9′), 3.90 (9H, s, 3-OCH3, 2′-OCH3, 6′-OCH3), 1.13 (3H, d, J = 6.4 Hz, H-9); 13C-NMR (125 MHz, CDCl3): δC 153.9 (C-2′, C-6′), 147.2 (C-3), 144.7 (C-4), 132.9 (C-1′), 132.0 (C-1). 131.1 (C-4′, C-7′), 128.5 (C-8′), 119.0 (C-6), 114.0 (C-5), 108.3 (C-2), 103.7 (C-3′, C-5′), 82.8 (C-8), 73.1 (C-7), 63.7 (C-9′), 56.5 (2′-OCH3, 6′-OCH3), 56.3 (3-OCH3), 12.8 (C-9).26
Myrifralignan F (14) – colorless oil, 1H-NMR (500 MHz, CDCl3): δH 6.68 (2H, s, H-2′, H-6′), 6.54 (2H, s, H-2, H-6), 6.58 (1H, d, J = 15.8 Hz, H-7′), 6.33 (1H, dt, J = 5.7, 15.8 Hz, H-8′), 4.78 (1H, d, J = 2.8 Hz, H-7), 4.34 (3H, overlapped, H-8, H-9′), 3.90, 3.87 (12H, s, -OCH3), 1.12 (3H, d, J = 6.4 Hz, H-9); 13C-NMR data, see reference.17
Myrifralignan E (15) – yellowish oil; ‒26.9 (c 0.02, CHCl3); 1H-NMR (500 MHz, CDCl3): δH 9.71 (1H, d, J = 7.6 Hz, H-9′), 7.43 (1H, d, J = 15.8 Hz, H-7′), 6.85 (2H, s, H-3′, H-5′), 6.67 (1H, dd, J = 7.6, 15.8 Hz, H-8′), 6.55 (2H, s, H-2, H-6), 4.78 (1H, d, J = 2.7 Hz, H-7), 4.43 (1H, qd, J = 2.7, 6.4 Hz, H-8), 3.94 (6H, s, 2′-OCH3, 6′-OCH3), 3.87 (6H, 3-OCH3, 5-OCH3), 1.15 (3H, d, J = 6.4 Hz, H-9); 13C-NMR (125 MHz, CDCl3): δC 193.4 (C-9′), 154.4 (C-2′, C-6′), 152.5 (C-7′), 147.1 (C-3, C-5), 138.0 (C-1′), 133.9 (C-4), 130.8 (C-1), 130.1 (C-4′), 128.4 (C-8′), 105.8 (C-3′, C-5′), 102.9 (C-2, C-6), 83.3 (C-8), 73.6 (C-7), 56.5 (2-OCH3, 6-OCH3, 2′-OCH3, 6′-OCH3), 12.9 (C-9).20
(7R,8R)-1-(4′-Hydroxy-3′-methoxyphenyl)-1-methoxy-2-(2″-methoxy-4″-(1‴-(E)-propenyl)-phenoxy) propane (16) – colorless oil, 1H and 13C NMR data, see reference.17
(+)-(4-Hydroxy-3-methoxy-l′-allyl-3′,5′-dimethoxy)-8-O-4′-neolignan (17) – yellow oil; ‒104.8 (c 0.02, CHCl3); 1H-NMR (500 MHz, CDCl3): δH 6.82 (1H, d, J = 8.0 Hz, H-5), 6.77 (1H, d, J = 1.8 Hz, H-2), 6.70 (1H, dd, J = 1.8, 8.0 Hz, H-6), 6.41 (2H, s, H-3′, H-5′), 5.97 (1H, ddt, J = 6.8, 10.0, 16.8 Hz, H-8′), 5.10 (2H, m, H-9′), 4.34 (1H, m, H-8), 3.85 (3H, s, 3-OCH3), 3.80 (6H, s, 2′-OCH3, 6′-OCH3), 3.34 (2H, d, J = 6.8 Hz, H-7′), 3.12 (1H, dd, J = 5.2, 13.5 Hz, H-7a), 2.73 (1H, dd, J = 8.2, 13.5 Hz, H-7b), 1.20 (3H, d, J = 6.2 Hz, H-9); 13C-NMR (125 MHz, CDCl3): δC 153.7 (C-2′, C-6′), 146.3 (C-3), 143.9 (C-4), 137.4 (C-8′), 135.5 (C-1′), 134.4 (C-1), 131.1 (C-4′), 122.2 (C-6′), 116.0 (C-9′), 114.1 (C-5), 112.3 (C-2), 105.7 (C-3′, C-5′), 80.1 (C-8), 56.1 (2′-OCH3, 6′-OCH3), 56.0 (3-OCH3), 43.0 (C-7), 40.6 (C-7′), 19.6 (C-9).27
Myrifralignan G (18) – colorless oil, 1H and 13C NMR data, see reference.17
Malabaricone B (19) – colorless amorphous solid; 1H-NMR (500 MHz, CD3OD): δH 7.18 (1H, t, J = 8.0 Hz, H-19), 6.96 (2H, d, J = 7.7 Hz, H-11, H-15), 6.68 (2H, d, J = 8.1 Hz, H-12, H-14), 6.34 (2H, d, J = 8.1 Hz, H-18, H-20), 3.10 (2H, t, J = 7.3 Hz, H-2), 2.47 (2H, t, J = 6.4 Hz, H-9), 1.65 (2H, d, J = 6.2 Hz, H-3), 1.53 (2H, m, H-8), 1.31 (8H, m, H-4, H-5, H-6, H-7); 13C-NMR (125 MHz, CD3OD): δC 209.7 (C-1), 163.3 (C-17, C-21), 156.0 (C-13), 136.8 (C-19), 134.9 (C-10), 130.2 (C-11, C-15), 116.0 (C-12, C-14), 111.4 (C-16), 108.4 (C-18, C-20), 45.7 (C-2), 36.0 (C-9), 32.9 (C-8), 30.5 (C-5, C-6, C-7), 30.2 (C-4), 25.7 (C-3).28
Malabaricone C (20) – colorless amorphous solid, 1H-NMR (500 MHz, CD3OD): δH 7.15 (1H, t, J = 8.2 Hz, H-19), 6.66 (1H, d, J = 8.0 Hz, H-14), 6.61 (1H, d, J = 2.0 Hz, H-11), 6.46 (1H, dd, J = 2.0, 8.0 Hz, H-15), 6.33 (2H, d, J = 8.2 Hz, H-18, H-20), 3.09 (2H, m, H-2), 2.41 (2H, m, H-9), 1.64 (2H, m, H-3), 1.51 (2H, m, H-8), 1.29 (8H, m, H-4, H-5, H-6, H-7); 13C-NMR (125 MHz, CD3OD): δC 209.6 (C-1), 163.3 (C-17, C-21), 145.8 (C-12), 143.8 (C-13), 136.8 (C-19), 135.8 (C-10), 120.6 (C-15), 116.4 (C-11), 116.1 (C-14), 111.3 (C-16), 108.3 (C-18, C-20), 45.7 (C-2), 36.2 (C-9), 32.8 (C-8), 30.5 (C-5, C-6, C-7), 30.2 (C-4), 25.6 (C-3).28
Myrifragranone A (21) – 1H and 13C NMR data, see reference.17
Myrifragranone B (22) – 1H and 13C NMR data, see reference.17
Myrifragranone C (23) – 1H-NMR (500 MHz, CD3OD): δH 6.97 (1H, d, J = 1.7 Hz, H-2), 6.69 (1H, d, J = 8.0 Hz, H-5), 6.75 (1H, dd, J = 8.0, 1.7 Hz, H-6), 4.46 (1H, d, J = 8.3 Hz, H-7), 5.18 (1H, dq, J = 12.4, 6.1 Hz, H-8), 1.08 (3H, d, J = 6.1 Hz, H-9), 6.37 (2H, s, H-3′, H-5′), 3.26 (2H, d, J = 6.6 Hz, H-7′), 5.92 (1H, m, H-8′), 5.04 (2H, m, H-9′), 3.77 (3H, s, OCH3), 3.71 (6H, s, OCH3), 3.10 (2H, m, H-2″), 1.64 (2H, m, H-3″), 1.30–2.42 (12H, m, H-3″–H-9″), 6.63 (1H, d, J = 1.9 Hz, H-11″), 6.69 (1H, d, J = 8.0 Hz, H-14″), 6.48 (1H, dd, J = 8.0, 1.9 Hz, H-15″), 6.34 (1H, d, J = 8.5 Hz, H-18″), 7.73 (1H, d, J = 8.5 Hz, H-19″); 13C NMR data, see reference.17
Myrifragranone D (24) – 1H and 13C NMR data, see reference.17
MTT assay for evaluating the cytotoxicity in ovarian cancer cell lines – TOV-112D and A2780 cells (cisplatin-sensitive ovarian carcinoma cell lines) were obtained from the American Type Culture Collection (VA, USA) and Dr. Stephen B. Howell (University of California, San Diego, USA), respectively. SK-OV3 cells (cisplatin-resistant ovarian carcinoma cell line) were supplied by Korean Cell Line Bank (Seoul, Korea). The cells were cultured under optimal conditions and subjected to mycoplasma detection weekly using e-Myco™ VALiD mycoplasma detection kit (iNtRON Biotechnology, Gyeonggi, Korea). MTT assay was used to determine the viability of the cells after 48 h of treatment with various concentrations of the test compound, according to the described procedure.29-34 The IC50 values were calculated using the online tool Quest Graph™ IC50 Calculator (AAT Bioquest, Inc., Pleasanton, CA, USA).
Western blotting – Protein expression was measured using western blotting as described previously.35 Whole-cell lysates were extracted using RIPA buffer (Pierce, Waltham, MA, USA), and protein concentrations were determined using BCA Protein Assay Kit (Pierce). Primary antibodies against p44/42 MAPK (p-ERK1/2) (#9101), p44/42 MAPK (ERK1/2) (#9102), phospho-p38 MAPK (Thr180/Tyr182) (p-p38 MAPK) (#9211), p38 MAPK (#9212), phospho-SAPK/JNK (Thr183/Tyr185) (p-SAPK/JNK) (#9251), SAPK/JNK (#9252), caspase-3 (#9662), and PARP (46D11) (#9532) were purchased from Cell Signaling Technology (Danvers, MA, USA). β‒actin (Santa Cruz Biotechnology, Dallas, TX, USA; sc-47778) was used as a loading control. The western blot bands were quantified using ImageJ.
Flow cytometric analysis – The percentage of cells undergoing active apoptosis was determined using FITC Annexin V Apoptosis Detection Kit I (BD Biosciences, Franklin Lakes, NJ, USA; #556547), following the manufacturer’s instructions. The cells were incubated in 6-well plates (5 × 105 cells/well) for 12 h. Then, they were treated with DMSO, 25 or 100 µM of the test compound for 48 h. Adherent and floating cells were collected, and two cold phosphate-buffered saline washes were performed, followed by FITC Annexin V and propidium iodide (PI) staining.36,37 Apoptosis was evaluated using a BD FACSAria™ III flow cytometer (BD Biosciences).
Statistical analysis – Data are expressed as the means ± standard deviations of three independent experiments. Student’s t-test was used to compare the two groups. All tests were two-sided, and P values of <0.05 were considered to indicate statistical significance.38
Results and Discussion
A phytochemical investigation on the methanolic extract of M. fragrans seeds was conducted and led to the isolation of 24 known compounds (1‒24). After analyzing NMR spectroscopic data, along with comparing the results with published literature, the structures of all compounds were successfully elucidated and presented in Fig. 1.
Structures of neolignans (1–18) and diarylnonanoid derivatives (19–24) isolated from the seeds of M. fragrans.
Compound (14) was obtained as a colorless oil. The NMR data of 14 showed signals of a neolignan moiety with aromatic protons at [δH 6.54 (2H, s, H-2, H-6), 6.68 (2H, s, H-3′, H-5′)], two oxygenated methine groups [δH 4.78 (1H, d, J = 2.8 Hz, H-7), 4.34 (1H, m, overlapped, H-8)], four methoxy groups [δH 3.90, 3.87 (12H, s)], and a methyl group [δH 1.12 (3H, d, J = 6.4 Hz, H-9)]. The configuration of a double bond at C-7′–C-8′ [δH 6.58 (1H, d, J = 15.8 Hz, H-7′), 6.33 (1H, dt, J = 15.8, 5.7 Hz, H-8′)] was identified as E-configuration by large coupling constant (J = 15.8 Hz).29 The configuration at C-7 and C-8 was determined as erythyo based on the coupling constant between H-7 and H-8 (J = 2.8 Hz) and the comparison of chemical shifts with published literature data.17 Consequently, compound 14 was elucidated as myrifralignan F.17
Compound 20 was obtained as a colorless amorphous solid. NMR data of 20 displayed signals of a diarylnonanoid with two substituted benzene rings [δH 6.61 (1H, d, J = 2.0 Hz, H-11), 6.66 (1H, d, J = 8.0 Hz, H-14), 6.46 (1H, dd, J = 8.0, 2.0 Hz, H-15), 7.15 (1H, t, J = 8.2 Hz, H-19), 6.33 (2H, d, J = 8.2 Hz, H-18, H-20)], a (CH2)8 group [δH 3.09 (2H, m, H-2), 1.64 (2H, m, H-3), 1.29 (8H, m, H-4, H-5, H-6, H-7), 1.51 (2H, m, H-8), 2.41 (2H, m, H-9)], and a carbonyl group [δC 209.6 (C-1)]. The linkage between aromatic rings through carbonyl and (CH2)8 groups was determined by HMBC correlations of H-2 with C-1 and C-16, H-8 with C-9 and C-10, and H-9 with C-10, C-11, and C-15. Thus, the structure of compound 20 was identified to be malabaricone C.28
Compound 23 was isolated as a yellow oil. The structure of 23 was comprised of two parts. Part A was determined as a neolignan with signals of two benzene rings [δH 6.97 (1H, d, J = 1.7 Hz, H-2), 6.69 (1H, d, J = 8.0 Hz, H-5), 6.75 (1H, dd, J = 1.7, 8.0 Hz, H-6), 6.37 (2H, s, H-3′, H-5′)], an oxygenated methine [δH 5.18 (1H, dq, J = 12.4, 6.1 Hz, H-8)], three methoxy groups [δH 3.77 (3H, s), 3.71 (6H, s)], and a vinyl group [δH 5.92 (1H, m, H-8′), 5.04 (2H, dq, J = 12.4, 6.1 Hz, H-9′)]. Whereas part B was determined as malabaricone C based on the analysis of NMR data.28 According to the above analysis and a comparison with data from literature, the structure of compound 23 was elucidated as myrifragranone C.17
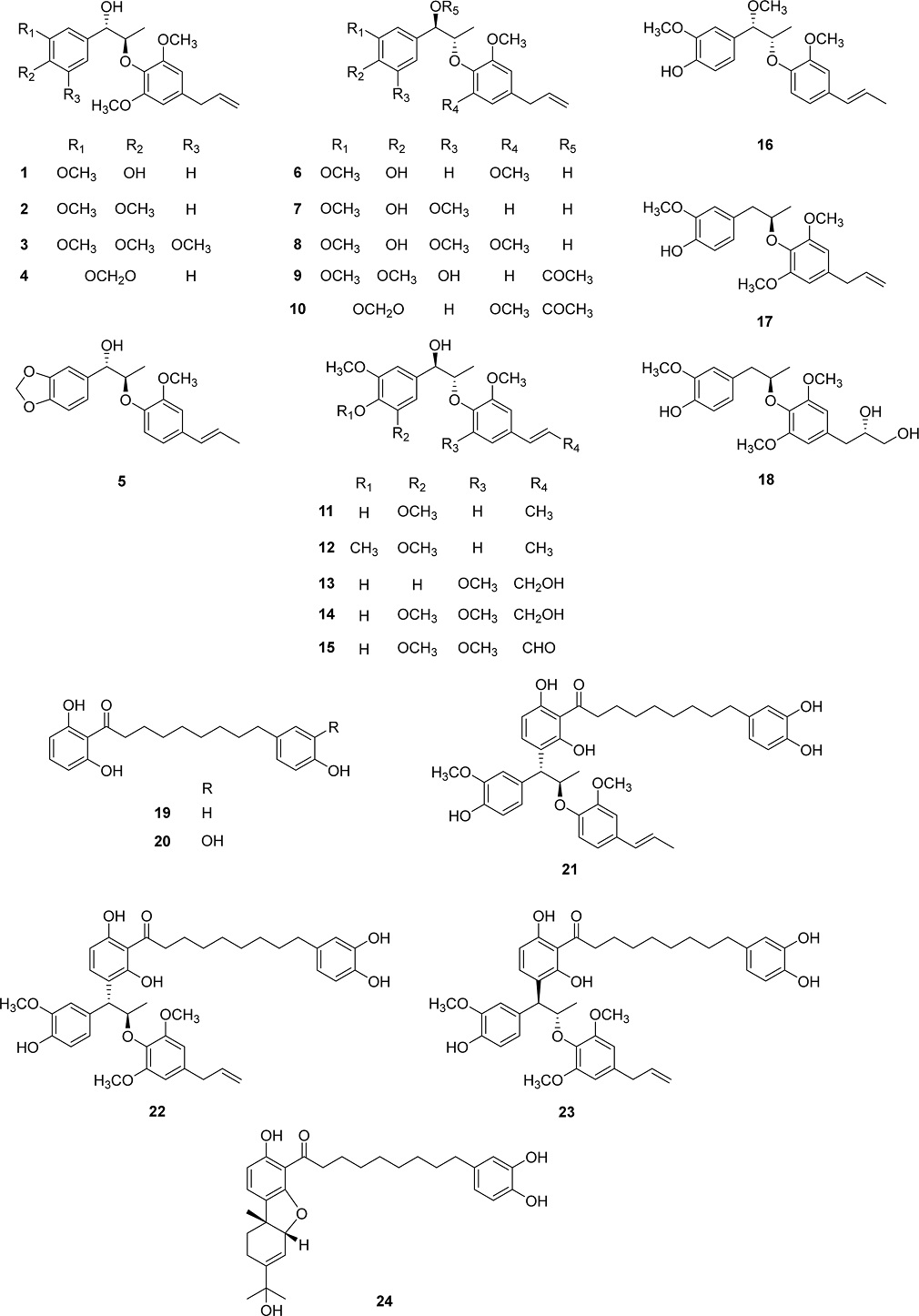
The cytotoxicity of all isolated compounds (1–24) in cisplatin-sensitive (A2780 and TOV-112D) and –resistant (SK-OV3) human ovarian cancer cell lines was evaluated using the MTT assay. Among them, neolignan myrifralignan F (14) exhibited moderate cytotoxicity against A2780, TOV-112D, and SK-OV3 cell lines with IC50 values of 27.3 ± 1.3, 45.3 ± 2.0, and 84.8 ± 3.3 μM, respectively (Fig. 2A). Previous studies reported that malabaricone C (20) exhibited cytotoxicity against MCF-7 human breast cancer and A549 lung cancer cell lines with IC50 values of 7.0 and 6.0 μM, respectively.39,40 Our results revealed that compound 20 also exhibited cytotoxicity against A2780, TOV-112D, and SK-OV3 cell lines with IC50 values of 36.1 ± 2.3, 48.9 ± 3.9, and 62.9 ± 4.8 μM, respectively (Fig. 2B). Meanwhile, myrifragranone C (23) with a diphenylnonanoid moiety similar to that of compound 20, which is linked to two phenylpropyl groups, displayed the most significant cytotoxicity among all isolated compounds, with IC50 values of 14.1 ± 1.1 (against A2780), 16.9 ± 1.1 (against TOV-112D), and 33.4 ± 1.3 (against SK-OV3) μM (Fig. 2C). The other compounds with a diphenylnonanoid moiety (compounds 21, 22, and 24) exhibited no cytotoxicity against all test cancer cell lines, suggesting that the absolute configuration 7S,8S in the chemical structure of diarylnonanoid derivatives affects their cytotoxicity. The remaining compounds had no effect on any of the test cancer cell lines.
Effects of compounds 14 (A), 20 (B), and 23 (C) on the viability of A2780, TOV-112D, and SK-OV3 cells detected by MTT assay. Data are presented as mean ± SD of three independent experiments.
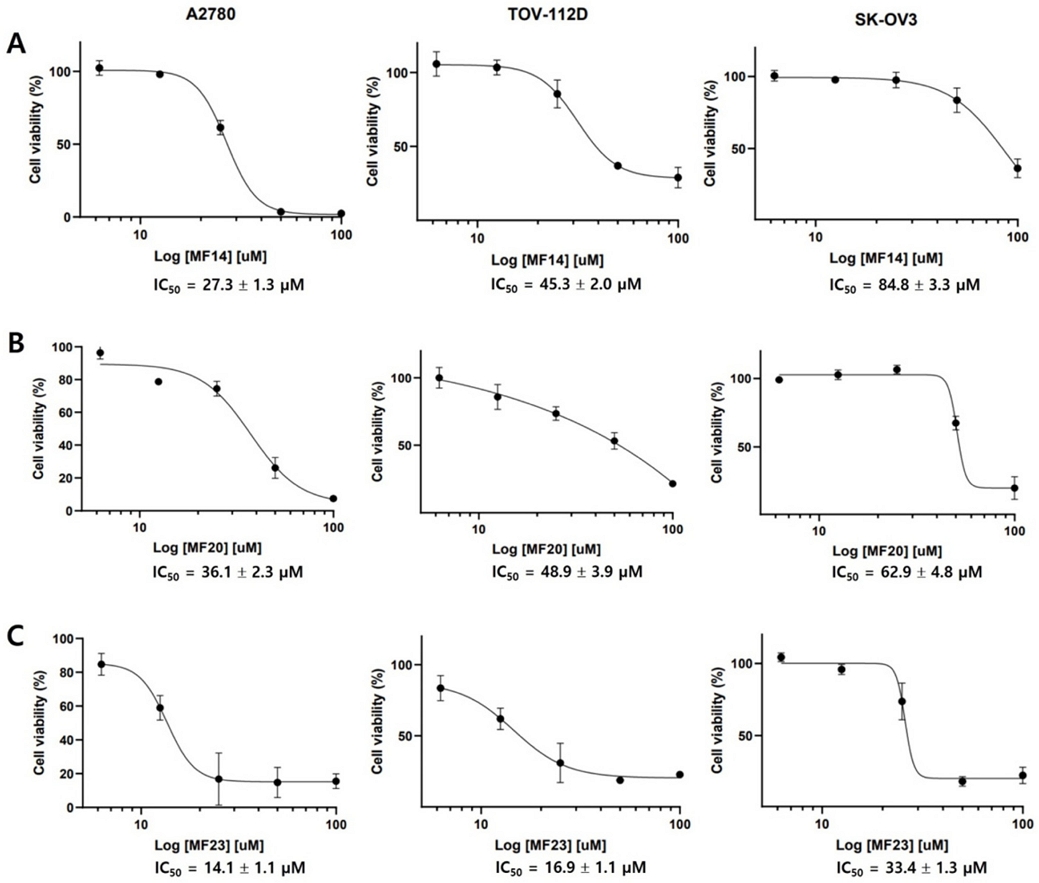
To investigate the mechanism underlying the cytotoxic effect of compound 23, human ovarian cancer cells treated with control or compound 23 (25 and 100 µM for A2780 and SK-OV3 cells, respectively) were stained with Annexin V and PI. In A2780 cells, compound 23 induced apoptosis after 48 h of treatment at a concentration of 25 μM with 7.9% comprising early apoptotic cells and 16.3% comprising late apoptotic cells, which increased 2‒3-fold compared with untreated control cells. Necrotic cells (4.1%) also increased compared with the control (1.3%) (Fig. 3). In SK-OV3 cells, compound 23 induced apoptosis at a concentration of 100 μM with 6.7% comprising early apoptotic cells and 21.5% comprising late apoptotic cells, equivalent to a 7‒9-fold increase compared with the controls (Fig. 3). Our results indicated that compound 23 significantly induced cell death through apoptosis in A2780 and SK-OV3 ovarian cancer cells.
Effect of compound 23-induced apoptosis in A2780 and SK-OV3 cells. FITC Annexin V apoptosis assay was used to examine the ratio of cellular apoptosis in A2780 and SK-OV3 cells after treatment with 25 μM or 100 μM of compound 23 for 48 h, respectively. Annexin V-positive cells ratio was shown in the bar graph. Data were obtained from three independent experiments. *P < 0.05, ***P < 0.001.
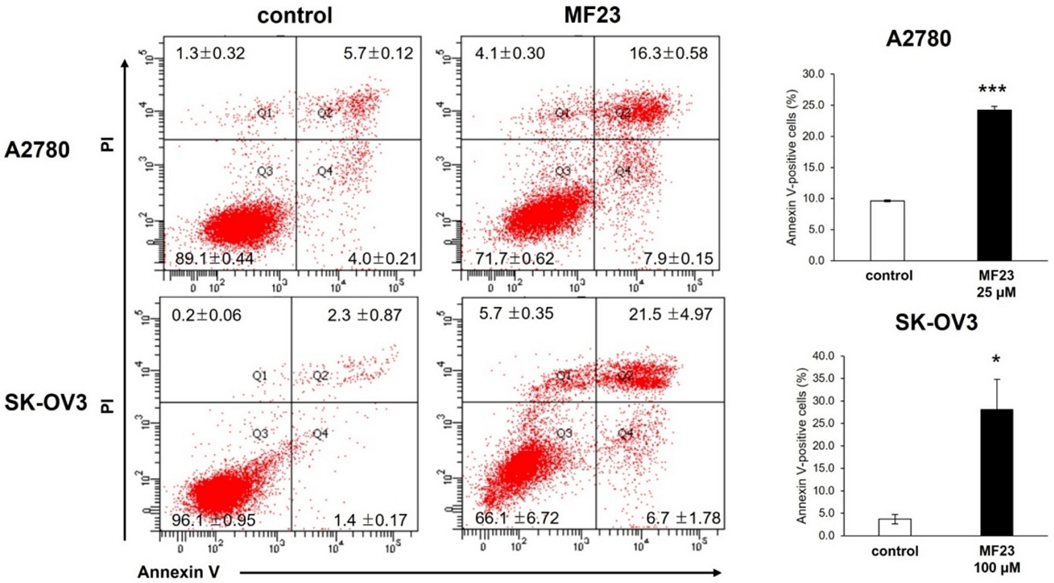
Caspases, also known as cysteine-aspartic proteases, are a family of protease enzymes required for initiating apoptosis within a cell.41 Caspase-3 is an effector caspase that is cleaved and consequently activated following the initiation of apoptosis.41 Cleaved caspase-3 transmits an apoptotic signal to downstream targets, including poly-ADP ribose polymerase (PARP) and other substrates, via enzymatic activity.42 To investigate the effect of compound 23 on cellular markers of apoptosis, cleaved caspase-3, and PARP levels were evaluated using western blotting, following the introduction of different concentrations of compound 23 to A2780 and SK-OV3 cells (25 and 100 µM for A2780 and SK-OV3 cells, respectively). As shown in Fig. 4, the levels of cleaved caspase-3 and PARP increased in compound 23-treated cells.
Effect of compound 23 on cellular markers of apoptosis. A2780 and SK-OV3 cells were subjected to control and 25 μM or 100 μM of compound 23, respectively. Apoptosis related markers were determined by western blotting analysis with specific antibodies caspase-3 and PARP. β-actin was used as a loading control.
Previous studies have revealed that MAPK pathways play a regulatory role in apoptosis.43,44 MAPK consists of three subfamilies, namely, extracellular signaling-regulated kinase (ERK), c-Jun N-terminal kinase (JNK), and p38, and is crucial for cellular functions.43 To determine the apoptosis-related signaling pathways activated in A2780 and SK-OV3 cells, the levels of pERK, pJNK, and pp38 were assessed using western blotting with different concentrations of compound 23 (25 and 100 µM for A2780 and SK-OV3 cells, respectively). The results revealed that compound 23 significantly increased the phosphorylation levels of JNK and p38 in both A2780 and SK-OV3 cells, whereas changes in the phosphorylation levels of ERK were different in the two cell lines (Fig. 5), suggesting that the activation of JNK and p38 is associated with compound 23-induced apoptosis.
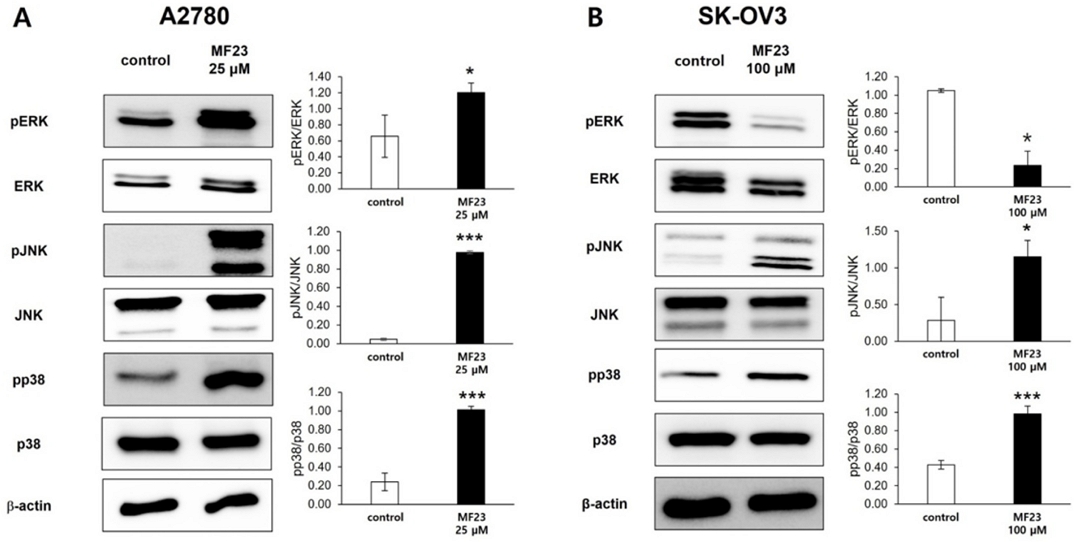
Compound 23-induced modulations of apoptosis-related signaling pathways in A2780 (A) and SK-OV3 (B) cells. A2780 and SK-OV3 cells were subjected to control and 25 μM or 100 μM of compound 23, respectively. Apoptosis-related signaling pathways were determined by western blotting analysis with specific antibodies as indicated. β-actin was used as a loading control. Data were obtained from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
In conclusion, 24 secondary metabolites, including 18 neolignans and 6 diarylnonanoid derivatives, were isolated from the seeds of M. fragrans. The cytotoxicity of all compounds against three human ovarian cancer cell lines (A2780, TOV-112D, and SK-OV3) was evaluated. Among them, myrifragranone C (23) exhibited the most potent cytotoxic effect on all test cancer cell lines, with IC50 values ranging from 14.1 to 33.4 µM. In addition, the mechanism of cytotoxicity induced by this active compound in ovarian cancer cells was performed. Our results reveal that compound 23 significantly induces cell death through apoptosis in A2780 and SK-OV3 ovarian cancer cells. Apoptosis is a crucial mechanism for maintaining cellular homeostasis, and the activation of caspase-3 is an important stage in initiating apoptosis. Compound 23 increased the levels of cleaved caspase-3 and PARP in treated cells, indicating the activation of the apoptotic pathway. Furthermore, the effect of compound 23 on MAPK pathways, which play a regulatory role in apoptosis, was also investigated. The results reveal that compound 23 significantly increases the phosphorylation levels of JNK and p38 in both A2780 and SK-OV3 cells, indicating the activation of the JNK and p38 pathways in compound 23-induced apoptosis. Our study provides insight into the underlying mechanism of the cytotoxic effect of compound 23 in ovarian cancer cells for the first time, which could lead to the development of promising therapies for ovarian cancer by targeting these pathways.
Acknowledgments
This research was supported by the National Research Foundation of Korea (NRF) funded by the Korea government (MSIT) (No. NRF-2020R1A5A2017323). The authors would like to thank Korea Basic Science Institute (KBSI) Daegu Center for the mass spectra measurement service.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
-
Huang, L.; Zhang, J.; Deng, Y.; Wang, H.; Zhao, P.; Zhao, G.; Zeng, W.; Wang, Y.; Chen, C.; Wagstaff, W.; Haydon, R. C.; Reid, R. R.; He, T. C.; Shen, L.; Luu, H. H.; Zhao, L. Genes Dis. 2023, 10, 1687-1701.
[https://doi.org/10.1016/j.gendis.2022.12.005]

-
Matulonis, U. A.; Sood, A. K.; Fallowfield, L.; Howitt, B. E.; Sehouli, J.; Karlan, B. Y. Nat. Rev. Dis. Primers 2016, 2, 16061.
[https://doi.org/10.1038/nrdp.2016.61]

-
Xu, X.; Chen, F.; Zhang, L.; Liu, L.; Zhang, C.; Zhang, Z.; Li, W. J. Ethnopharmacol. 2021, 279, 114343.
[https://doi.org/10.1016/j.jep.2021.114343]

-
Henri, J. L.; Nakhjavani, M.; McCoombe, S.; Shigdar, S. Biochimie 2023, 204, 108-117.
[https://doi.org/10.1016/j.biochi.2022.09.005]

-
Mikuła-Pietrasik, J.; Witucka, A.; Pakuła, M.; Uruski, P.; Begier-Krasińska, B.; Niklas, A.; Tykarski, A.; Książek, K. Cell. Mol. Life Sci. 2019, 76, 681-697.
[https://doi.org/10.1007/s00018-018-2954-1]

-
Anand, U.; Dey, A.; Chandel, A. K. S.; Sanyal, R.; Mishra, A.; Pandey, D. K.; De Falco, V.; Upadhyay, A.; Kandimalla, R.; Chaudhary, A.; Dhanjal, J. K.; Dewanjee, S.; Vallamkondu, J.; Pérez de la Lastra, J. M. Genes Dis. 2022, 10, 1367-1401.
[https://doi.org/10.1016/j.gendis.2022.02.007]

-
McWhinney, S. R.; Goldberg, R. M.; McLeod, H. L. Mol. Cancer Ther. 2009, 8, 10-16.
[https://doi.org/10.1158/1535-7163.MCT-08-0840]

-
Guastalla 3rd, J. P.; Diéras, V. Br. J. Cancer 2003, 89, S16-S22.
[https://doi.org/10.1038/sj.bjc.6601496]

-
Trang, N. M.; Kim, E. N.; Lee, H. S.; Jeong, G. S. Biomolecules 2022, 12, 256.
[https://doi.org/10.3390/biom12020256]

-
Van Cong, P.; Anh, H. L. T.; Trung, N. Q.; Quang Minh, B.; Viet Duc, N.; Van Dan, N.; Trang, N. M.; Phong, N. V.; Vinh, L. B.; Anh, L. T.; Lee, K. Y. Nat. Prod. Res. 2023, 37, 2342-2350.
[https://doi.org/10.1080/14786419.2022.2042534]

-
Rastegari, A.; Manayi, A.; Rezakazemi, M.; Eftekhari, M.; Khanavi, M.; Akbarzadeh, T.; Saeedi, M. BMC Chem. 2022, 16, 106.
[https://doi.org/10.1186/s13065-022-00897-9]

-
Ha, M. T.; Vu, N. K.; Tran, T. H.; Kim, J. A.; Woo, M. H.; Min, B. S. Arch. Pharm. Res. 2020, 43, 1067-1092.
[https://doi.org/10.1007/s12272-020-01285-4]

-
Patro, B. S.; Bauri, A. K.; Mishra, S.; Chattopadhyay, S. J. Agric. Food Chem. 2005, 53, 6912-6918.
[https://doi.org/10.1021/jf050861x]

-
Li, C.-W.; Chu, Y.-C.; Huang, C.-Y.; Fu, S.-L.; Chen, J.-J. Molecules 2020, 25, 5198.
[https://doi.org/10.3390/molecules25215198]

-
Sathya, S.; Amarasinghe, N. R.; Jayasinghe, L.; Araya, H.; Fujimoto, Y. South Afr. J. Bot. 2020, 130, 172-176.
[https://doi.org/10.1016/j.sajb.2019.12.020]

-
Rathee, J. S.; Patro, B. S.; Brown, L.; Chattopadhyay, S. Free Radic. Res. 2016, 50, 111-121.
[https://doi.org/10.3109/10715762.2015.1112005]

-
Oanh, V. T.; Phong, N. V.; Min, B. S.; Yang, S. Y.; Kim, J. A. J. Enzyme Inhib. Med. Chem. 2023, 38, 2251099.
[https://doi.org/10.1080/14756366.2023.2251099]

-
Li, F.; Yang, X. W. Phytochemistry 2008, 69, 765-771.
[https://doi.org/10.1016/j.phytochem.2007.09.008]

-
Duan, L.; Tao, H. W.; Hao, X. J.; Gu, Q. Q.; Zhu, W. M. Planta Med. 2009, 75, 1241-1245.
[https://doi.org/10.1055/s-0029-1185506]

-
Cao, G. Y.; Xu, W.; Yang, X. W.; Gonzalez, F. J.; Li, F. Food Chem. 2015, 173, 231-237.
[https://doi.org/10.1016/j.foodchem.2014.09.170]

-
Zacchino, S. A.; Badano, H. J. Nat. Prod. 1988, 51, 1261-1265.
[https://doi.org/10.1021/np50060a037]

-
Rye, C. E.; Barker, D. Eur. J. Med. Chem. 2013, 60, 240-248.
[https://doi.org/10.1016/j.ejmech.2012.12.013]

-
Hattori, M.; Hada, S.; Shu, Y. Z.; Kakiuchi, N.; Namba, T. Chem. Pharm. Bull. 1987, 35, 668-674.
[https://doi.org/10.1248/cpb.35.668]

-
Francis, S. K.; James, B.; Varughese, S.; Nair, M. S. Nat. Prod. Res. 2019, 33, 1204-1208.
[https://doi.org/10.1080/14786419.2018.1457670]

-
Morikawa, T.; Hachiman, I.; Matsuo, K.; Nishida, E.; Ninomiya, K.; Hayakawa, T.; Yoshie, O.; Muraoka, O.; Nakayama, T. J. Nat. Prod. 2016, 79, 2005-2013.
[https://doi.org/10.1021/acs.jnatprod.6b00262]

-
Olajide, O. A.; Ajayi, F. F.; Ekhelar, A. I.; Awe, S. O.; Makinde, J. M.; Alada, A. R. Phytother. Res. 1999, 13, 344-345.
[https://doi.org/10.1002/(SICI)1099-1573(199906)13:4<344::AID-PTR436>3.0.CO;2-E]

-
Kasahara, H.; Miyazawa, M.; Kameoka, H. Phytochemistry 1995, 38, 343-346.
[https://doi.org/10.1016/0031-9422(94)00643-8]

-
Kundu, K.; Nayak, S. K. J. Nat. Prod. 2017, 80, 1776-1782.
[https://doi.org/10.1021/acs.jnatprod.6b01119]

-
Phong, N. V.; Anh, D. T. N.; Chae, H. Y.; Yang, S. Y.; Kwon, M. J.; Min, B. S.; Kim, J. A. Bioorg. Chem. 2022, 128, 106072.
[https://doi.org/10.1016/j.bioorg.2022.106072]

-
Trang, N. M.; Kim, E. N.; Pham, T. H.; Jeong, G. S. J. Agric. Food Chem. 2023, 71, 10037-10049.
[https://doi.org/10.1021/acs.jafc.3c00368]

-
Huong, P. T. M.; Phong, N. V.; Huong, N. T.; Trang, D. T.; Thao, D. T.; Cuong, N. X.; Nam, N. H.; Van Thanh, N. J. Nat. Med. 2022, 76, 210-219.
[https://doi.org/10.1007/s11418-021-01582-2]

- Retnowati, R.; Sulistyarti, H.; Wahidah, N. Z.; Syarifah, A. L.; Salam, S.; Nurlelasari.; Safari, A.; Harneti, D.; Tanjung, M.; Hidayat, A. T.; Maharani, R.; Supratman, U.; Shiono, Y. Nat. Prod. Sci. 2021, 27, 18-27.
- Shehu, A.; Ponnapalli, M. G.; Mahboob, M.; Prabhakar, P. V.; Olatunji, G. A. Nat. Prod. Sci. 2022, 28, 62-67.
-
Chung, H. S. Nat. Prod. Sci. 2022, 28, 115-120.
[https://doi.org/10.20307/nps.2022.28.3.115]

-
Kim, E. N.; Trang, N. M.; Kang, H.; Kim, K. H.; Jeong, G. S. Cells 2022, 11, 3596.
[https://doi.org/10.3390/cells11223596]

-
Vinh, L. B.; Phong, N. V.; Ali, I.; Dan, G.; Koh, Y. S.; Anh, H. L. T.; Van Anh, D. T.; Yang, S. Y.; Kim, Y. H. Med. Chem. Res. 2020, 29, 2020-2027.
[https://doi.org/10.1007/s00044-020-02613-5]

-
Huong, P. T. M.; Phong, N. V.; Thao, N. P.; Binh, P. T.; Thao, D. T.; Thanh, N. V.; Cuong, N. X.; Nam, N. H.; Thung, D. C.; Minh, C. V. J. Asian Nat. Prod. Res. 2020, 22, 193-200.
[https://doi.org/10.1080/10286020.2018.1543280]

-
Vu, N. K.; Le, T. T.; Woo, M. H.; Min, B. S. Nat. Prod. Sci. 2021, 27, 176-182.
[https://doi.org/10.20307/nps.2021.27.3.176]

-
Tyagi, M.; Patro, B. S.; Chattopadhyay, S. Free Radic. Res. 2014, 48, 466-477.
[https://doi.org/10.3109/10715762.2014.886328]

-
Tyagi, M.; Bauri, A. K.; Chattopadhyay, S.; Patro, B. S. Free Radic. Biol. Med. 2020, 148, 182-199.
[https://doi.org/10.1016/j.freeradbiomed.2020.01.011]

-
Julien, O.; Wells, J. A. Cell Death Differ. 2017, 24, 1380-1389.
[https://doi.org/10.1038/cdd.2017.44]

-
Mashimo, M.; Onishi, M.; Uno, A.; Tanimichi, A.; Nobeyama, A.; Mori, M.; Yamada, S.; Negi, S.; Bu, X.; Kato, J.; Moss, J.; Sanada, N.; Kizu, R.; Fujii, T. J. Biol. Chem. 2021, 296, 100046.
[https://doi.org/10.1074/jbc.RA120.014479]

-
Roux, P. P.; Blenis, J. Microbiol. Mol. Biol. Rev. 2004, 68, 320-344.
[https://doi.org/10.1128/MMBR.68.2.320-344.2004]

- Kang, F. C.; Chen, Y. C.; Wang, S. C.; So, E. C.; Huang, B. M. Int. J. Mol. Med. 2020, 46, 439-448.