NO Production Inhibition of Lignans from Vietnamese Schisandra sphenanthera Rehd. et Wils. Fruits
Abstract
In the present study, bioactivity-guided extraction and isolation of the n-hexane fraction of the fruits of Vietnamese Schisandra sphenanthera led to the isolation of five dibenzocyclooctadiene lignans as gomisin N (1), schisandrin C (2), gomisin H (3), gomisin D (4), and gomisin C (5). All the isolates were tested for their inhibition of NO production in LPS-induced RAW 264.7 cells. Among them, compounds 4 and 5 showed weak inhibition of NO production with IC50 values of 25.0 ± 1.6 and 24.8 ± 2.0 µM, respectively. Compound 1 exhibited NO production inhibition with an IC50 value of 15.8 ± 2.1 µM, meanwhile, schisandrin C (2) showed the most potent inhibition with an IC50 value of 8.5 ± 0.5 µM. In addition, compound 2 had a concentration-dependent inhibitory effect on the protein expression of the inflammatory enzymes iNOS and COX-2. Their physicochemical properties and ADMET data were predicted by in silico, indicating favorable drug-like properties as well as low acute oral toxicity. The results suggest that the fruit of S. sphenanthera and its phytochemical constituents might be used as anti-inflammatory agents.
Keywords:
Schisandra sphenanthera, NO production, Lipopolysaccharide, RAW 264.7 macrophage cell lineIntroduction
Inflammation is a protective, whole-body biological reaction against inflammatory agents such as microorganisms, chemical and physical agents, or endogenous agents such as localized tissue necrosis due to ischemia, and autoimmune diseases.1 The symptoms of inflammation usually seen at the site include swelling, heat, redness, and pain. The body’s inflammatory condition can be divided into acute and chronic inflammation.1,2 Acute inflammation occurs in response to sudden inflammatory agent attacks in a short period, while longer-acting inflammatory agents prompt chronic inflammation.3,4 Many inflammatory mediators participate in the regulation of the inflammatory reaction. These include amino acids, arachidonic acid metabolites (prostaglandins, leukotrienes, and lipoxins), cytokines (interleukin-1β, interleukin-6β, and tumor necrosis factor-α), platelet-activating factors, neuropeptides, and nitric oxide, all of which are released from cells.5-7 In addition, inflammatory mediators are generated by supplementary systems: the kinin system, coagulation system, and fibrinolytic system.8-10 Among these mediators, nitric oxide (NO) is produced by enzyme synthesis (via nitric oxide synthase, NOS) which is induced (iNOS) in macrophages and other cells involved in the inflammatory response.11-13 A large amount of NO can stimulate many important proteins and enzymes for inflammatory reactions, such as NF-κB.14 To study anti-inflammatory agents, the model of macrophage RAW 264.7 cells induced by lipopolysaccharides (LPS) is used.13,14 Macrophages are involved in chronic inflammatory reactions by producing different inflammatory mediators, including cytokines, chemokines, interferons, bacterial stimulation factors, lysozymes, proteases, growth factors, eicosanoids, and NO.13,14 Among these, NO is overproduced by the inflammatory enzyme iNOS, thus leading to various diseases, including asthma, arthritis, multiple sclerosis, colitis, psoriasis, neurodegenerative disorders, tumor growth, and graft rejection due to septic shock.12 Therefore, finding new NO-production inhibitors for anti-inflammatory treatment is necessary.
Schisandra sphenanthera Rehd. et Wils. is a member of the Schisandraceae plant family.15 The plant can be found primarily in China, North America, Russia, South Korea, and Japan.16,17 S. sphenanthera was discovered on Ngoc Linh Mountain in Quang Nam province, central Vietnam, and was classified as a new species for Vietnam in 2006.18 The species was listed as “Endangered in Vietnam” on the Vietnam Medicinal Plant Red List in 2006 due to its limited range, minimal natural reserves, and diminished habitat due to deforestation around Ngoc Linh Mountain.18 Some research groups are now working on generating a supply of genetic material for medicine production by propagating S. sphenanthera and developing cultivation areas for it in the mountainous region of Quang Nam province.18 S. sphenanthera is used in a similar way to S. chinensis, which is a dried fruit that is imported and widely used in traditional Chinese medicine for the treatment of many diseases such as hepatitis, cough, coronary heart disease, dizziness, tinnitus, and asthenia due to its important medicinal properties.16,19,20 Like S. chinensis, S. sphenanthera has a sour and bitter taste and a warming property and affects the lungs and kidneys. It has the effect of calming the lungs, relieving coughs, stopping sweating, promoting kidney function, and generating bodily fluids.16,20 Many studies have been conducted on the biological activities of S. sphenanthera worldwide, including on its ability to induce cancer cell apoptosis,18-20 and on its anti-inflammatory, antioxidant properties, anti-coronary heart disease properties, and treatment of Alzheimer's disease.15,21-25 To date, more than 25 plant species in the Schisandraceae family have been studied for their chemical composition, with many compounds from different chemical groups isolated, including lignans and triterpenoids.16,17,20 However, there are very few studies on S. sphenanthera collected in Vietnam. In this study, we report the extraction and structural identification of compounds from an n-hexane fraction of S. sphenanthera, and their anti-inflammatory activity. In silico assays are also applied for better understanding the anti-inflammatory mechanism as well as the ADMET properties of potential compounds.
Experimental
Plant materials - Fresh S. sphenanthera fruit samples were harvested in Ngoc Linh Mountain area, Quang Nam province in October 2021 and identified by Mr. Nguyen Xuan Phai, from the Traditional Medicine Association in Da Nang city. The identified sample has the code TMH-2019-NVTN and is stored in the Laboratory of Pharmacognosy and Quality Control, School of Medicine and Pharmacy, The University of Danang.
Preparation of crude extract - S. sphenanthera fruits were dried at 45oC to constant weight. Two kg of dried fruit were extracted with ethanol 80% three times using an extractor pot, each time for 3 hours. The extracts were combined and filtered through a coarse filter paper. Then the extract was evaporated at 45oC to obtain 350 g of the total extract. Dissolve 340 g in 3 liters of warm water, and then successively partitioned with n-hexane, ethyl acetate, and n-butanol, to obtain corresponding fractionated extracts of n-hexane (50 g), ethyl acetate (24 g), n-butanol (105 g), and a remaining aqueous solution. The n-hexane fraction extract (48 g) was subjected to column chromatography on silica gel and eluted with a linear solvent system of n-hexane:acetone from 100:1 to 1:1 (v/v), yielding ten subfractions (H-1 to H-10) based on the analysis of the results on the TLC sample. Subfraction H-2 (2.1 g) was further purified on a silica gel column using n-hexane: EtOAc (60:1, v/v) as the eluent, resulting in five smaller fractions, H-2.1 to H-2.5. Compound 1 (125 mg) was obtained as a white powder precipitate from fraction H-2.2. Fraction H-3 (1.1 g) was subjected to column chromatography on silica gel and eluted with a solvent system of n-hexane:acetone (40:1 to 25:1, v/v), yielding eight subfractions from H-3.1 to H-3.8. Compound 2 (14 mg) was obtained as a precipitate from fraction H-3.3. Subfraction H-4 (1.2 g) was subjected to column chromatography on silica gel and eluted with a solvent system of n-hexane:ethyl acetate (30:1 to 10:1, v/v), yielding twelve fractions from H-4.1 to H-4.12. Compound 3 (25 mg) was obtained from fraction H-4.1 as a precipitate in a mixture of n-hexane:acetone (10:1). Fraction H-4.2 (1.5 g) was further purified on a silica gel column using a linear solvent system of n-hexane:ethyl acetate (25:1 to 10:1, v/v), yielding 4 (28 mg) and fraction H-4.2.1. Compound 5 (14 mg) was obtained as a precipitate after overnight standing of fraction H-4.2.1.
Gomisin N (1) - 1H-NMR (500 MHz, DMSO-d6): δH (ppm) 6.70 (1H, s, H-4), 2.61 (1H, dd, J = 13.5, 7.7 Hz, H-6a), 2.80 (1H, dd, J = 13.5, 2.2 Hz, H-6b), 1.86 (1H, m, H-7), 1.75 (1H, m, H-8), 2.20 (1H, dd, J = 13.2, 9.5 Hz, H-9a), 2.43 (1H, dd, J = 13.5, 1.9 Hz, H-9b), 6.55 (1H, s, H-11), 5.96 (2H, d, J = 1.3 Hz, O–CH2–O), 3.45 (3H, s, 1-OCH3), 3.78 (3H, s, 2-OCH3), 3.82 (3H, s, 3-OCH3), 3.84 (3H, s, 14-OCH3), 0.75 (3H, d, J = 7.2 Hz, H-17), 0.98 (3H, d, J = 7.2 Hz, H-18); 13C-NMR (125 MHz, DMSO-d6): δC (ppm) 151.5 (C-1), 140.2 (C-2), 152.0 (C-3), 110.8 (C-4), 134.5 (C-5), 38.5 (C-6), 33.6 (C-7), 40.9 (C-8), 35.0 (C-9), 137.7 (C-10), 102.0 (C-11), 148.5 (C-12), 133.6 (C-13), 141.0 (C-14), 121.5 (C-15), 123.4 (C-16), 21.3 (C-17), 11.9 (C-18), 60.2 (1-OCH3), 59.5 (2-OCH3), 55.6 (3-OCH3), 58.4 (14-OCH3), 101.0 (O–CH2–O).
Schisandrin C (2) - 1H-NMR (500 MHz, DMSO-d6): δH (ppm) 6.58 (1H, s, H-4), 2.54 (1H, dd, J = 13.5, 7.4 Hz, H-6a), 2.27 (1H, dd, J = 13.5, 7.2 Hz, H-6b), 1.64 (1H, m, H-7), 1.75 (1H, m, H-8), 1.95 (1H, d, J = 13.0 Hz, H-9a), 2.22 (1H, dd, J = 13.0, 7.2 Hz, H-9b), 6.57 (1H, s, H-11), 5.99 (2H, brd, J = 1.3 Hz, 2-O–CH2–O-3), 5.99 (2H, brd, J = 1.3 Hz, 12-O–CH2–O-13), 3.71 (3H, s, 1-OCH3), 3.72 (3H, s, 14-OCH3), 0.65 (3H, d, J = 7.2 Hz, H-17), 0.93 (3H, d, J = 7.2 Hz, H-18); 13C-NMR (125 MHz, DMSO-d6): δC (ppm) 140.6 (C-1), 134.3 (C-2), 147.0 (C-3), 105.5 (C-4), 132.0 (C-5), 39.3 (C-6), 32.2 (C-7), 40.2 (C-8), 34.6 (C-9), 137.4 (C-10), 102.0 (C-11), 148.5 (C-12), 134.2 (C-13), 140.0 (C-14), 120.7 (C-15), 121.8 (C-16), 21.0 (C-17), 12.5 (C-18), 58.9 (1-OCH3), 59.0 (14-OCH3), 100.8 (2-O–CH2–O-3), 100.8 (12-O–CH2–O-13).
Gomisin H (3) - 1H-NMR (500 MHz, DMSO-d6): δH (ppm) 6.90 (1H, s, H-4), 2.65 (1H, d, J = 13.4 Hz, H-6a), 2.30 (1H, d, J = 13.7 Hz, H-6b), 1.85 (1H, m, H-8), 2.45 (1H, dd, J = 14.2, 8.3 Hz, H-9a), 2.90 (1H, dd, J = 14.2, 2.2 Hz, H-9b), 6.75 (1H, s, H-11), 3.76 (3H, s, 1-OCH3), 3.68 (3H, s, 2-OCH3), 3.78 (3H, s, 3-OCH3), 3.90 (3H, s, 12-OCH3), 3.85 (3H, s, 13-OCH3), 1.72 (3H, d, J = 7.2 Hz, H-17), 1.16 (3H, s, H-18); 13C-NMR (125 MHz, DMSO-d6): δC (ppm) 154.1 (C-1), 141.2 (C-2), 153.3 (C-3), 112.8 (C-4), 136.5 (C-5), 42.9 (C-6), 70.3 (C-7), 36.4 (C-8), 21.3 (C-9), 138.0 (C-10), 114.4 (C-11), 153.0 (C-12), 136.3 (C-13), 141.8 (C-14), 124.0 (C-15), 129.5 (C-16), 36.4 (C-17), 16.2 (C-18), 61.1 (1-OCH3), 61.5 (2-OCH3), 57.1 (3-OCH3), 56.9 (12-OCH3), 61.4 (13-OCH3).
Gomisin D (4) - 1H-NMR (500 MHz, DMSO-d6): δH (ppm) 6.85 (1H, s, H-4), 6.10 (1H, s, H-6), 1.82 (1H, m, H-8), 2.65 (1H, dd, J = 14.0, 8.0 Hz, H-9a), 1.90 (1H, brd, J = 14.0 Hz, H-9b), 6.62 (1H, s, H-11), 1.42 (3H, d, J = 8.0 Hz, H-17), 1.25 (3H, s, H-18), 3.53 (3H, s, 1-OCH3), 3.80 (3H, s, 2-OCH3), 3.91 (3H, s, 3-OCH3), 5.84 (2H, brs, 12-O–CH2–O-13), 2.10 (1H, m, H-3’), 3.54 (1H, m, H-4’a), 3.62 (1H, m, H-4’b), 1.06 (3H, s, H-5’), 1.18 (3H, d, J = 7.2 Hz, H-6’); 13C-NMR (125 MHz, DMSO-d6): C (ppm) 152.3 (C-1), 138.2 (C-2), 152.0 (C-3), 112.5 (C-4), 132.7 (C-5), 87.0 (C-6), 71.5 (C-7), 45.2 (C-8), 37.0 (C-9), 137.2 (C-10), 103.5 (C-11), 149.2 (C-12), 138.6 (C-13), 143.7 (C-14), 122.6 (C-15), 124.5 (C-16), 28.0 (C-17), 19.5 (C-18), 61.4 (1-OCH3), 61.1 (2-OCH3), 57.6 (3-OCH3), 101.8 (12-O–CH2–O-13), 177.5 (C-1’), 76.5 (C-2’), 39.0 (C-3’), 74.0 (C-4’), 25.1 (C-5’), 12.0 (C-6’).
Gomisin C (5) - 1H-NMR (500 MHz, DMSO-d6): δH (ppm) 6.94 (1H, s, H-4), 4.77 (1H, d, J = 6.0 Hz, H-6), 2.35 (1H, m, H-7), 1.92 (1H, m, H-8), 2.45 (1H, dd, J = 13.0, 9.0 Hz, H-9a), 2.12 (1H, d, J = 13.0 Hz, H-9b), 6.67 (1H, s, H-11), 5.85 (2H, d, J = 1.3 Hz, O–CH2–O), 3.74 (3H, s, 1-OCH3), 3.82 (3H, s, 2-OCH3), 3.76 (3H, s, 3-OCH3), 3.85 (3H, s, 14-OCH3), 1.06 (3H, d, J = 7.0 Hz, H-17), 1.22 (3H, s, H-18), 7.30–7.57 (H2’-H-6’); 13C-NMR (125 MHz, DMSO-d6): δC (ppm) 150.4 (C-1), 140.1 (C-2), 151.6 (C-3), 111.5 (C-4), 134.2 (C-5), 79.5 (C-6), 40.1 (C-7), 42.8 (C-8), 34.5 (C-9), 135.4 (C-10), 103.1 (C-11), 105.2 (C-12), 134.0 (C-13), 141.1 (C-14), 121.6 (C-15), 122.5 (C-16), 28.1 (C-17), 18.4 (C-18), 60.4 (1-OCH3), 60.5 (2-OCH3), 55.8 (3-OCH3), 58.6 (14-OCH3), 148.4 (O–CH2–O), 166.0 (C-1’), 133.6 (C-2’), 128.2 (C-3’), 122.0 (C-4’), 132.3 (C-5’), 120.5 (C-6’), 127.8 (C-7’).
Materials for anti-inflammatory bioactivity assay method - Lipopolysaccharides (LPS) from Escherichia coli from Sigma Chemical Co. (St. Louis, MO, USA). Dulbecco’s Modified Eagle’s Medium (DMEM), fetal bovine serum (FBS) from Life Technologies, Inc., (Gaithersburg, MD, USA). Sodium nitrite, sulfanilamide, N-1-napthylethylenediamine dihydrochloride, and dimethyl sulfoxide (DMSO) were purchased from Sigma Chemical Co. (St. Louis, MO, USA). Other necessary chemicals were purchased from GIBCO, Invitrogen, and Promega.
Cell culture –The RAW264.7 cell line was cultured in DMEM medium with 2 mM L-glutamine, 10 mM HEPES, and 1.0 mM sodium pyruvate, supplemented with 10% fetal bovine serum (FBS) (GIBCO). Cells were cultured after 3-5 days with the ratio (1:3) and incubated in a CO2 incubator at 37oC, 5% CO2.
NO production in RAW264.7 macrophage cells - Incubation of RAW 274.7 cells in 96-well plates at 37oC and 5% CO2 for 24 h was performed. DMEM medium without FBS was replaced with the culture medium for three hours, then cells were incubated at different concentrations for two hours before LPS (10 g/mL) stimulated NO factor production for 24 hours. NG-methyl-L-arginine acetate (L-NMMA) (Sigma) at concentrations of 100; 20; 4 and 0.8 g/mL were used as positive controls, whereas as negative controls only sample diluent was injected into the wells, while the negative control was sample diluent alone. The determination of nitrite (NO2-), one of the indicators of NO generation, will be carried out using the Griess Reagent System (Promega Corporation, WI, USA). 100 mL of cell culture medium was transferred to a new 96-well plate and added Griess reagent: 50 liters of 1% sulfanilamide in 5% phosphoric acid and 50 liters of 0.1% N-1-naphthylethylenediamine dihydrochloride. A 540 nm microplate reader was used to measure nitrite content after 10 minutes of room temperature incubation. As a blank well, non-FBS DMEM medium was used, and the nitrite content of each sample of the test was determined using the NaNO2 standard concentration curve, and then compared against the negative control. As a result of the following formula, it was determined that the sample had the ability to inhibit NO production in the sample:
- % inhibition = 100% - (Nosample content /NoLPS content) * 100
The value of IC50 (concentration that inhibits 50% of the nitrogen oxide formation) was calculated using the TableCurve 2Dv4 computer software.
Western Blotting analysis - RAW 264.7 cells were washed with PBS, incubated and lysed with buffer solution [10% glycerol, 1% Triton X-100, 1 mM Na3PO4, 1 mM EGTA, 10 mM NaF, 1 mM Na4P2O7, 20 mM Tris buffer (pH 7.9), 100 mM β-glycerophosphate, 137 mM NaCl, 5 mM EDTA] for about 1 h in ice. Then, centrifuge at 12000 rpm for 30 minutes at 4oC and wash the protein several times with PBS. Dissolve the protein in cold PBS in preparation for gel electrophoresis.26,27 Carry out loading of 20-30 μg protein, separate the proteins by electrophoresis using 10% SDS-PAGE. After electrophoresis, transfer the separated proteins on the gel to a polyvinylidene difluoride membrane. After transferring from the gel to the membrane, the proteins on the membrane are inactivated with 5% skim milk at room temperature in Tris-buffer saline−Tween solution (TBST; 20 mM Tris, 500 mM NaCl, pH 7.5, 0.1% Tween 20). The membrane is then incubated with monoclonal anti-iNOS or anti-COX2 (diluted 1:1000) in 5% non-fat milk/TBST for 2 h at room temperature. After incubation with the primary antibody, the membrane is washed 3 times with TBST at room temperature and then incubated with IgG secondary antibody (antimouse IgG secondary antibody, Sigma, St. Louis, MO, USA) diluted 1:2000 in 2.5% non-fat milk/TBST for 1 h at room temperature. The membrane containing the protein is washed 3 times with TBST and the immune protein reaction is detected by chemiluminescence-ECL (Amersham International PLC, Buckinghamshire, UK) using hyperfilm and chemiluminescent reagents. The Western blot results are quantified by measuring the optical density correlation with the control sample using Fujifilm Image Reader Las-4000 software (FujiFilm Corp., Tokyo, Japan).
Protein and ligand preparation - The crystal structure of iNOS, and COX-2 were downloaded from RSCB Protein Data Bank (PDB ID: 3E7G, and 5IKQ, respectively). All non-standard residues were removed using UCSF Chimera 1.17.3. Polar hydrogen atoms and Gasteiger charges are added to the proteins by “Dock Prep” tool of UCSF Chimera. The PubChem CID of two compounds (158103, and 119112, respectively) are pasted to the “Build Structure” tool of UCSF Chimera for ligand formation (ligands 1 and 2). Ligand preparation is done by “Dock Prep” tool of UCSF Chimera where hydrogen atoms are added and Gasteiger charges are assigned to the ligands. The energy minimisation of the ligands is executed by the “Minimize Structure” tool of UCSF Chimera. The prepared proteins and ligands are saved in PDB format.
Molecular docking - In this study, AutoDock Vina 1.2.5 was used for the molecular docking process and integrated into UCSF Chimera. CASTp 3.0 server28 was applied to investigate the active site of the proteins. To cover the active biding sites, the grids were centred at the area that includes all the residues pointed out by CASTp. The best conformers were searched based on Broyden-Fletcher-Goldfarb-Shanno algorithm. For each ligand, the number of conformers was set at maximum of 10 during the molecular docking process. Default parameters of AutoDock Vina were selected for the docking performance. After the docking process, the conformers were ranked according to their binding free energy with the proteins, in which the selection was done on the least binding free energy among all generated conformers. All the AutoDock Vina docking performances were run under Windows 10 Pro operating system, on the processor of 2.53 GHz Intel Core i5.
ADMET prediction - Gomisin N (1) and schisandrin C (2) were screened for the prediction of their physicochemical properties based on the “Lipinski’s rule of five”.29 In this process, SwissADME30 was employed to calculate some pharmacokinetic-related data of two compounds. Acute oral toxicity data was generated by DL-AOT Prediction Server.31
Result and Discussion
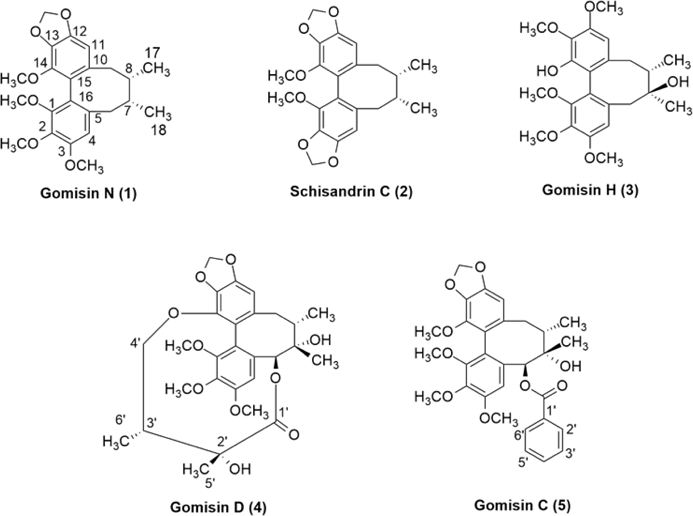
The 80% ethanolic extract was successively partitioned with n-hexane, ethyl acetate, and n-butanol, to obtain corresponding fractionated extracts of n-hexane, ethyl acetate, n-butanol (105 g), and a remaining aqueous solution. The n-hexane fraction extract was subjected to column chromatography on silica gel to obtain five natural compounds (1–5) (Fig. 1).
Compound 1 was obtained as a white powder. Under thin-layer chromatography with the solvent system n-hexane:acetone (85:15), 1 did not show any spots under visible light conditions, but showed spots in the UV-Vis region, and circular spots were observed in 20% vanillin–H2SO4 solution. The 1H-NMR spectrum of 1 showed signals at δH 6.70 (1H, s, H-4) and 6.55 (1H, s, H-11), equivalent to two protons attached to the aromatic ring. The signals at δH 2.59 (1H, dd, J = 13.5, 7.7 Hz, H-6α), 2.80 (1H, dd, J = 13.3, 2.2 Hz, H-6β), δH 2.20 (1H, dd, J = 13.2, 9.5 Hz, H-9α), and 2.43 (1H, dd, J = 13.5, 1.9 Hz, H-9β) were predicted to belong to two methylene groups. The two methine proton signals at δH 1.86 (1H, m) and 1.75 (1H, m, H-8), were observed and assigned to H-7 and H-8, respectively. The proton signal at δH 5.96 (d, 2H, J = 1.3 Hz) indicated the presence of a dioxymethylene group. The peaks at δH 3.45 (3H, s), 3.78 (3H, s), 3.82 (3H, s), and 3.84 (3H, s) were predicted to be four methoxy groups attached to the aromatic rings. In addition, the peaks at δH 0.75 (d, 3H, J = 7.2 Hz) and 0.98 (3H, d, J = 7.2 Hz) were determined to be two methyl (-CH3) groups. The 13C-NMR spectrum showed that compound 1 contains 23 carbons, including 12 carbons with signals in the aromatic region, such as δC 151.5 (C-1), 140.2 (C-2), 152.0 (C-3), 110.8 (C-4), 134.5 (C-5), 137.7 (C-10), 102.0 (C-11), 148.5 (C-12), 133.6 (C-13), 141.0 (C-14), 121.5 (C-15), and 123.4 (C-16), indicating that 1 also contains two aromatic rings in its structure. This information is typical of dibenzocyclooctadiene lignan skeletons in other Schisandra species.18,20,22 The peaks at δC 60.2, 59.5, 55.6, and 58.4 corresponded to the four methoxy (-OCH3) groups that were in accordance with the 1H-NMR spectrum. The peaks at δC 38.5 and 35.0 were determined to be carbons in the two methyl groups at positions C-6 and C-9 (-CH2-), and the peak at δC 101.0 was assigned to the dioxymethylene group (-OCH2O-). The remaining δC positions were 12.0 (7-CH3), 21.3 (8-CH3), and two methine carbons at δC 33.6 (C-7) and 40.9 (C-8). Based on the analysis of the 1H and 13C spectra above, this compound was identified as a dibenzocyclooctadiene lignan skeleton, and named gomisin N.16,20
Compound 2 was obtained in the form of white powder. The 1H-NMR spectrum of 2 showed two signals at δH 6.58 (2H, s), which are equivalent to two protons attached to the aromatic ring; compared to 1, these two protons were determined to be located at C-4 and C-11. The signals at δH 2.54 (d, 1H, J = 7.45 Hz) and 2.27 (dd, 1H, J = 13.4, J = 7.2 Hz) indicated that they are two hydrogens in the same methylene at H-6. The proton signals at δH 1.95 (d, 1H, J = 13.0 Hz) and δH 2.22 (dd, 1H, J = 13.0, 7.2 Hz) also indicated that these two protons are two hydrogens in another methylene group at H-9. At δH 1.64 (1H, m) and δH 1.75 (1H, m) were two protons of two methine groups that may be attached to a substituent with multiple hydrogens; they were therefore split into multiple peaks, similar to those of H-7 and H-8 of 1. The signal appearing at δH 5.99 (2H, brd) was a broad signal due to the splitting of 2H, which corresponded to the appearance of two dioxymethyl (-O-CH2-O-) groups with overlapping peaks. In addition, at δH 3.0–4.5 ppm, there were signals at δH 3.71 (3H, s) and 3.72 (3H, s) indicating two methoxy (-OCH3) groups, and at δH 0.93 (d, 3H, J = 7.2) and δH 0.65 (d, 3H, J = 7.2 Hz) there were two methyl (-CH3) groups in the formula. The 13C-NMR spectrum indicated signals for 22 carbons in the compound, including 12 carbons with signals in the aromatic region, as in 1. The appearance of signals at δC 58.9 and 59.0 was due to the presence of methoxy (-OCH3) groups. In addition, the signal at δC 100.8 was due to the presence of two dioxymethyl (-OCH2O-) groups. Similarly, to 1, the signals at δC 12.5, 21.0, 32.2, 34.6, 39.4, and 40.2 corresponded to (-CH-), (-CH2-), and (-CH3) groups, respectively. From the 1H and 13C spectra, we predict that 2 is also a dibenzocyclooctadiene lignan like 1; however, 2 differs from 1 in that it has an additional dioxymethyl group attached to an aromatic ring16,20. When compared to other lignan structures extracted from S. sphenanthera, the 1H- and 13C-NMR spectra of 2 are consistent with all positions of schisandrin C.32
Compound 3 was obtained in the form of white powder that dissolved well in acetone. The 1H-NMR spectrum of 3 shows signals at δH 6.90 (s, 1H, H-4) and 6.73 (s, 1H, H-11) that are equivalent to two protons attached to the aromatic ring at the same positions as compounds 1 and 2. Two pairs of proton signals-at positions δH 2.65 (d, 1H, J = 13.4 Hz) and 2.30 (d, 1H, J = 13.7 Hz), and δH 2.45 (dd, 1H, 14.2, J = 8.3 Hz) and 2.90 (dd, 1H, J = 14.7, 2.2 Hz)-were identified as two pairs of two methylene groups at positions H-6 and H-9 in the cyclooctadiene framework of the lignan skeleton. The current signal at δH 1.85 (m, 1H) could be assigned to a methine, while the five singlet signals at δH 3.76 (3H, s), 3.68 (3H, s), 3.78 (3H, s), 3.90 (3H, s), and 3.85 (3H, s) indicated that there were five methoxy groups (-OCH3). Interestingly, there was a singlet signal at δH 1.16 (3H, s, 7-CH3), indicative of the presence of a methyl group attached to the quaternary carbon. The other methyl group, at δH 1.72 (d, 3H, J = 7.2 Hz), could be localized to 8-CH3 of the cyclooctadiene ring. The 13C-NMR spectrum shows signals for 23 carbons in the formula, including 12 carbons of two aromatic rings. The carbon signals at δC 73.1, 61.5, 61.4, 56.9, and 57.1 corresponded to the five methoxy (-OCH3) groups. The peak at δC 73.0 corresponded to an oxygenated carbon signal at C-7. In addition, signals at δC 16.2, 16.9, 21.3, 36.4, and 42.9 corresponded to (-CH-), (-CH2-), and (-CH3) groups. From the above 1H- and 13C-NMR spectra, and based on comparison with compounds 1 and 2, compound 3 was shown to have the same cyclooctadiene lignan skeleton. However, 3 was distinguished by the presence of two OH groups at positions C-14 of the aromatic ring and C-7 of the cyclooctane ring, which is consistent with the compound named gomisin H.33
Compound 4 was obtained as a white powder. Under thin layer chromatography with the solvent system n-hexane:acetone (85:15), 4 did not show any spots under visible light, but appeared in the UV-Vis region and was observed as a round spot in 20% vanillin–H2SO4 solution. The 1H-NMR spectrum of 4 also showed the presence of two proton signals of the aromatic ring at δH 6.85 (1H, s, H-4) and 6.52 (1H, s, H-11). Three methoxy groups at δH 3.53 (3H, s, 1-OCH3), 3.80 (3H, s, 2-OCH3), and 3.91 (3H, s, 3-OCH3), and two signals for the dioxymethylene (O-CH2-O) group at δH 5.85 (2H, brs) were observed. The proton signals at δH 1.18 (3H, d, J = 7.2 Hz, H-6’), 1.06 (3H, s, H-5’), 1.25 (3H, s, H-18), and 1.42 (3H, d, J =8.0 H-17) were assigned to four methyl groups. In addition, two signals for the methine group were found at δH 1.82 (1H, m, H-8) and 2.10 (1H, m, H-3’). The remaining signals at δH 3.54 (1H, m, H-4’a), 3.62 (1H, m, H-4’b), and 2.65 (1H, dd, J = 14.0, 8.0 Hz, H-9a), 1.90 (1H, brd, J = 14.0 Hz, H-9b) indicated the presence of the two methylene groups. The 13C-NMR spectrum of 4 showed the presence of 27 carbon signals, including 12 signals in the aromatic region, indicating that the compound also contains two aromatic rings. Three signals were observed for the methoxy groups attached to the aromatic ring at δC 61.4 (1-OCH3), 61.1 (2-OCH3), and 57.6 (3-OCH3). There was one dioxymethylene carbon signal (δC 101.8) and two oxygenated carbons, of which one belonged to a quaternary carbon at δC 71.5 (C-7), and the other was a tertiary carbon at δC 87.0 (C-6). Compared to the other dibenzocyclooctadiene lignans,33 this compound had an additional side chain connecting C-6 to C-14 of the dibenzocyclooctadiene skeleton. This side chain was confirmed by the presence of a carbonyl carbon at δC 177.5 (C-1’), an oxygenated quaternary carbon at δC 76.5 (C-2’), a methine at δC 39.0 (C-3’), an oxygenated methylene at δC 74.0 (C-4’), and methyl groups at δC 25.1 (C-5’) and 12.0 (C-6’). Comparison of the 1H- and 13C-NMR spectra of 4 with the literature led to the identification of this compound as gomisin D.34
Compound 5 was obtained as white and acetone-soluble powder. Under thin layer chromatography with the solvent system n-hexane:acetone (85:15), 5 did not show any spots under visible light conditions but showed up as spots in the UV-Vis region, and was visible with a 20% vanillin–H2SO4 solution. The 1H-NMR spectrum of 5 showed the presence of signals from two protons on the aromatic ring at δH 6.94 (1H, s, H-4) and 6.67 (1H, s, H-11). There were four methoxy group signals at δH 3.74 (3H, s, 1-OCH3), 3.82 (3H, s, 2-OCH3), 3.76 (3H, s, 3-OCH3), and 3.85 (3H, s, 14-OCH3). This compound also contained a dioxymethylene group at δH 5.85 (2H, s). The 1H-NMR spectra of 5 also showed the presence of two methyl signals at δH 1.06 (3H, d, J = 7.0 Hz, H-17) and 1.22 (3H, s, H-18). One methine signal at δH 1.84 (1H, t, J = 7.9, 8.65, 16.05, H-8) was observed. Further methylene-group signals were observed at δH 2.45 (1H, dd, J = 13.0, 9.0 Hz, H-9a), and 2.12 (1H, d, J = 13.0 Hz, H-9b). Interestingly, this compound exhibited the presence of five more aromatic proton signals on the side chain, at δH 7.30–7.57 ppm. The 13C-NMR spectrum of 5 showed the presence of 30 carbon signals. Among them, 22 carbon signals were quite similar to the main dibenzocyclooctadiene lignan skeleton.33,34 However, this compound differed from the others by its additional benzoic acid moiety at δC 166.0 (C-1’), 133.6 (C-2’), 128.2 (C-3’), 122.0 (C-4’), 132.3 (C-5’), 120.5 (C-6’), and 127.8 (C-7’) (Fig. 1). Comparison with the 1H and 13C-NMR spectra of 5 with the literature indicated that this compound was consistent with the positions of gomisin C.35
To test the cytotoxic effects of compounds 1–5 on RAW 264.7 cells, we used the MTT method (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) on cell groups with and without LPS. Results showed that compounds 1–5 did not affect cell viability even at high concentrations of 50 μM after 24 h of incubation. To test the compounds’ inhibitory activity on NO production, RAW 264.7 cells were treated with isolated compounds at a concentration range of 0–30 μM, and the level of NO production after LPS-induced inflammation was determined by the nitrite concentration on the surface of the cells. Compound 3 showed weak inhibitory activity on NO production at concentrations higher than 30 μM and was considered inactive. Meanwhile, compounds 4 and 5 inhibited NO production, with IC50 values of 25.0 ± 1.6 and 24.8 ± 2.0 μM, respectively. Compound 1 exhibited inhibitory effect with an IC50 value of 15.8 ± 2.1 μM, whereas compound 2 had the strongest inhibitory activity with an IC50 value of 8.5 ± 0.5 μM. In this experiment, celastrol, a secondary metabolite, was used as a positive control.36 Celastrol inhibited NO production, with an IC50 value of 1.03 ± 0.12 μM.
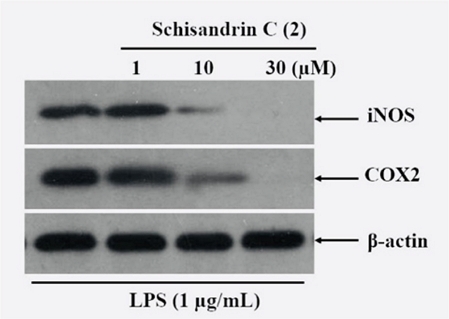
iNOS is one of the three main enzymes that produce NO from the amino acid L-arginine. NO from iNOS plays an important role in many physiological manifestations of diseases, such as in blood-pressure regulation, inflammation, infection, and malignant diseases.11,12 iNOS has been studied as a marker and therapeutic target in these diseases, especially inflammation. Cyclooxygenase-2 (COX-2), on the other hand, is the enzyme responsible for producing inflammatory mediators such as prostaglandins (PG) and their metabolites, such as PGE2, PGF2α, and PGD2.37 Since compound 2 showed the strongest inhibitory activity on NO production in RAW 264.7 cells during inflammation, it was selected to test the inhibitory effects on iNOS and COX-2 using western blotting analysis. The results showed that the protein concentrations of both iNOS and COX-2 were not detected when the cells were not stimulated with LPS (Fig. 2). However, when cells were stimulated with LPS, both cytokines were significantly upregulated in terms of their protein concentrations (Fig. 2). Interestingly, when the concentration of 2 was increased from 0 to 30 μM, the protein concentrations of both iNOS and COX-2 gradually decreased in LPS-stimulated cells. This indicates that 2 had a concentration-dependent inhibitory effect on the expression of inflammatory enzymes. In this experiment, the expression of alpha-tubulin protein did not change under all conditions tested.
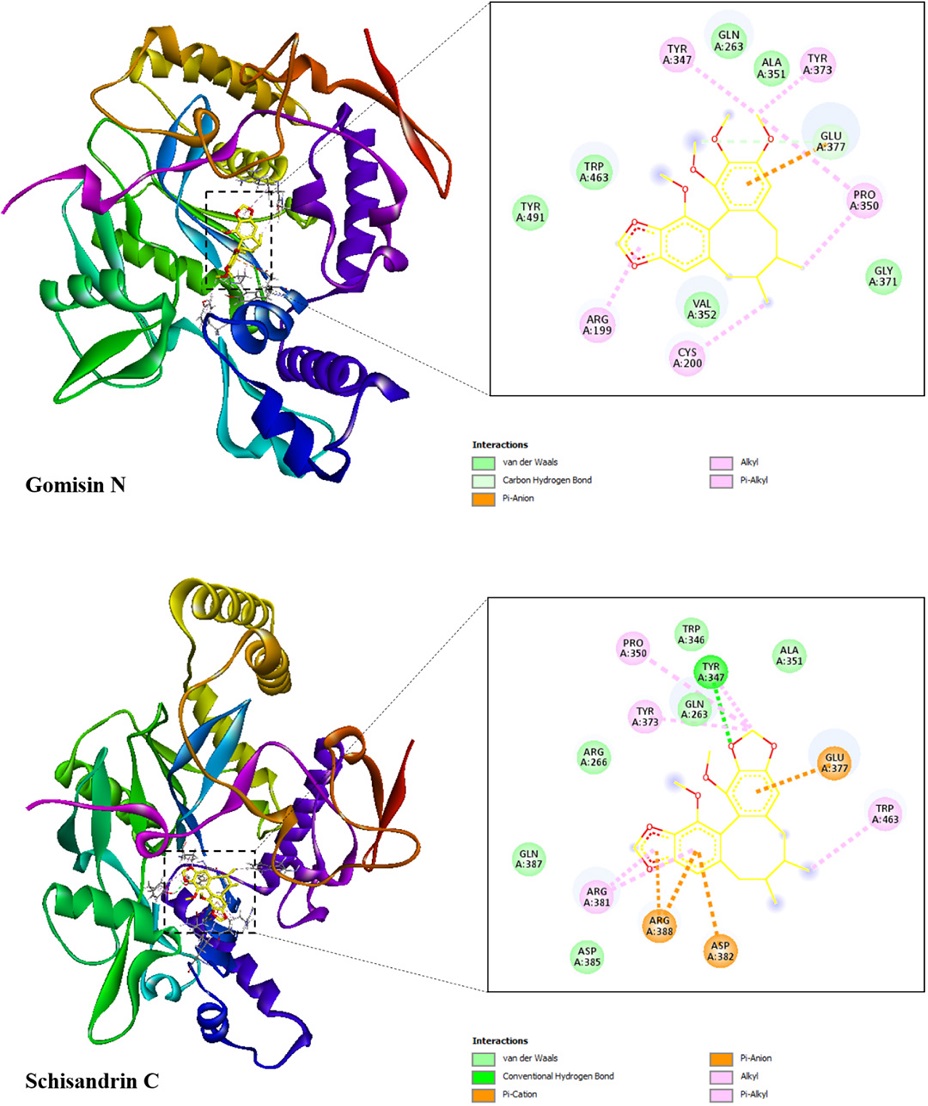
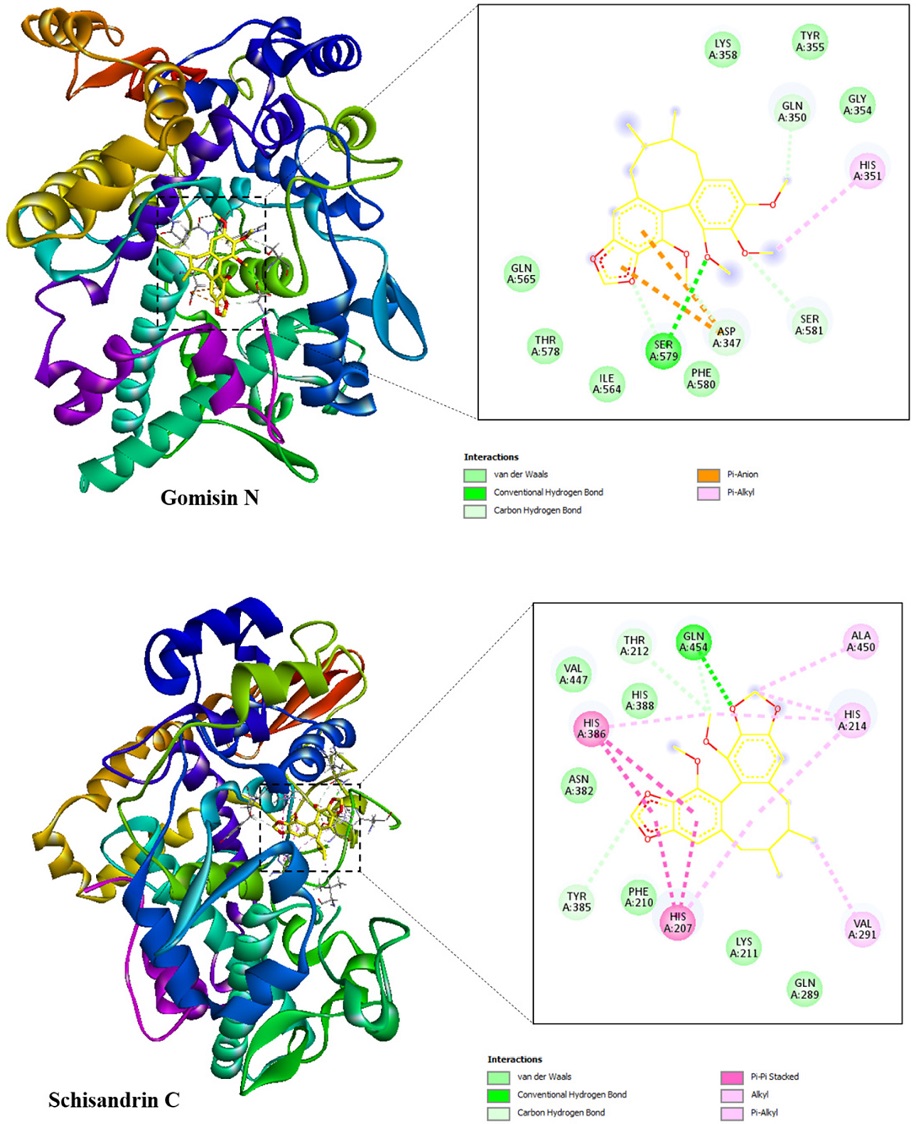
Molecular docking was used to investigate the interactions between two compounds (1 and 2) and the proteins of iNOS and COX-2 since these compounds showed good results in the anti-inflammatory assay. The docking results toward iNOS and COX-2 proteins including binding free energy, hydrogen bond, and hydrophobic interaction were illustrated in Table 2 and Table 3, respectively. The best docked poses of the ligands were exhibited in Fig. 3 and Fig. 4, showing their interactions with the iNOS and COX-2 targets, respectively.
Two potential compounds were suggested to demonstrate strong affinity toward the iNOS protein as their binding free energies were determined to be –7.2 and –8.4 kcal/mol for gomisin N (1), and schisandrin C (2), respectively. Gomisin N (1) interacted with iNOS protein through one hydrogen bond, which was located at Glu377. This ligand formed six hydrophobic interactions with Arg199, Cys200, Tyr347, Pro350, Tyr373, and Glu377. In case of schisandrin C (2), this ligand formed one hydrogen bond with Tyr347 along with eight hydrophobic interactions with the residues of Tyr347, Pro350, Tyr373, Glu377, Arg381, Asp382, Arg388, and Trp463.
The binding free energy of gomisin N (1), and schisandrin C (2) in complex with COX-2 receptor were –6.9 and –8.0 kcal/mol, respectively. Gomisin N (1) formed four hydrogen bonds with Asp347, Gln350, Ser579, and Ser581 along with two hydrophobic interactions with Asp347, and His351. Its counterpart, schisandrin C (2), interacted with COX-2 protein through three hydrogen bonds, which were located at Thr212, Tyr385, and Gln454. This ligand formed five hydrophobic interactions with the residues of His207, His214, Val291, His386, and Ala450.
The classic “Lipinski's Rule of Five” has traditionally served as a criterion for assessing a compound's drugability. In this study, both gomisin N (1) and schisandrin C (2) have molecular weights under 500 Daltons. They also exhibit fewer than 5 hydrogen bond donors, fewer than 10 hydrogen bond acceptors, and log P values below 5. These criteria indicate compliance with Lipinski's rule, suggesting favorable drug-like properties for these compounds (Table 4). Additionally, we evaluated the number of rotatable bonds, total polar surface area (TPSA), and aqueous solubility (log S) as physicochemical parameters. To ensure good oral bioavailability and intestinal absorption, the number of rotatable bonds should not exceed 10, and the TPSA value should stay below 140 Ų. Gomisin N (1) has 4 rotatable bonds, and schisandrin C (2) has 2, both well within the acceptable range. They share a TPSA value of 55.38 Ų, which is also favorable. Their log S values of –5.60 and –5.56, respectively, indicate moderate solubility. Comprehensive data on these compounds are provided in Table 4. Furthermore, Table 5 presents in silico predictions of the ADME (Absorption, Distribution, Metabolism, and Excretion) properties of gomisin N (1) and schisandrin C (2). These compounds were predicted to exhibit high gastrointestinal absorption and the ability to permeate the blood-brain barrier. Several cytochrome P enzymes play a crucial role in drug biotransformation, including CYP1A2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4. Gomisin N (1) and schisandrin C (2) were predicted to inhibit CYP2C19, CYP2C9, and CYP2D6 but not CYP3A4. They are also not expected to be substrates of P-glycoprotein (P-gp). The LD50 values of gomisin N (1) and schisandrin C (2) were calculated using the DL-AOT Prediction Server and found to be 3.17 and 3.27, respectively (Table 6). Based on these predicted results, these compounds fall into the “None required” group, indicating their potential use as therapeutic drugs.
In conclusion, we successfully isolated five natural compounds as gomisin N (1), schisandrin C (2), gomisin H (3), gomisin D (4) and gomisin C (5) from the n-hexane fraction of Vietnamese S. sphenanthera fruit. Their structures were elucidated using 1H–NMR and 13C-NMR spectroscopy, and comparisons were made with compounds previously obtained from the same plant. Among these compounds, schisandrin C demonstrated the most potent inhibitory activity with the lowest IC50 value against NO synthesis in LPS-stimulated RAW 264.7 cells. This effect was further validated through the reduction of iNOS and COX-2 enzyme protein concentrations. Molecular docking studies also indicated a strong binding affinity of gomisin N (1), schisandrin C (2) to the iNOS and COX-2 proteins. Additionally, predictions of their physicochemical properties and ADMET data suggested favorable drug-like characteristics and low acute oral toxicity. These collective findings point towards the potential of the n-hexane extract of S. sphenanthera fruit and its constituents as a valuable natural source for developing therapeutic agents targeting inflammation-related diseases.
Acknowledgments
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Conflict of interest
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
-
Abraham, C.; Cho, J. H. N. Engl. J. Med. 2009, 361, 2066–2078.
[https://doi.org/10.1056/NEJMra0804647]

-
Thia, K. T.; Loftus, E. V.; Sandborn, W. J.; Yang, S. K. Am. J. Gastroenterol. 2008, 103, 3167–3182.
[https://doi.org/10.1111/j.1572-0241.2008.02158.x]

-
Karlinger, K.; Györke, T.; Makö, E.; Mester, Á.; Tarján, Z. Eur. J. Radiol. 2000, 35, 154–167.
[https://doi.org/10.1016/S0720-048X(00)00238-2]

-
Mak, W. Y.; Zhao, M.; Ng, S. C.; Burisch, J. J. Gastroenterol. Hepatol. 2020, 35, 380–389.
[https://doi.org/10.1111/jgh.14872]

-
Hanna, A.; Frangogiannis, N. G. Cardiovasc. Drugs Ther. 2020, 34, 849–863.
[https://doi.org/10.1007/s10557-020-07071-0]

-
Gonçalves Dos Santos, G.; Delay, L.; Yaksh, T. L.; Corr, M. Front. Immunol. 2020, 10, 3061.
[https://doi.org/10.3389/fimmu.2019.03061]

-
Fattori, V.; Hohmann, M. S. N.; Rossaneis, A. C.; Manchope, M. F.; Alves-Filho, J. C.; Cunha, T. M.; Cunha, F. Q.; Verri, W. A. Jr. Expert. Opin. Ther. Targets 2017, 21, 1141–1152.
[https://doi.org/10.1080/14728222.2017.1398734]

-
Vanderwall, A. G.; Milligan, E. D. Front. Immunol. 2019, 10, 3009.
[https://doi.org/10.3389/fimmu.2019.03009]

-
Fujisawa, S.; Konnai, S.; Okagawa, T.; Maekawa, N.; Tanaka, A.; Suzuki, Y.; Murata, S.; Ohashi, K. BMC Vet. Res. 2019, 15, 68.
[https://doi.org/10.1186/s12917-019-1813-0]

-
Li, P.; Zheng, Y.; Chen, X. Front. Pharmacol. 2017, 8, 460.
[https://doi.org/10.3389/fphar.2017.00460]

-
Abu-Amara, M.; Yang, S. Y.; Seifalian, A.; Davidson, B.; Fuller, B. Liver Int. 2012, 32, 531–543.
[https://doi.org/10.1111/j.1478-3231.2012.02755.x]

-
Miyake, T.; Yokoyama, Y.; Kokuryo, T.; Mizutani, T.; Imamura, A.; Nagino, M. J. Surg. Res. 2013, 183, 742–751.
[https://doi.org/10.1016/j.jss.2013.01.048]

-
González, C.; Herradón, E.; Abalo, R.; Vera, G.; Pérez-Nievas, B. G.; Leza, J. C.; Martín, M. I.; López-Miranda, V. Diabetes Metab. Res. Rev. 2011, 27, 331–340.
[https://doi.org/10.1002/dmrr.1176]

-
Zhang, Y. Q.; Ding, N.; Zeng, Y. F.; Xiang, Y. Y.; Yang, M. W.; Hong, F. F.; Yang, S. L. World J. Gastroenterol. 2017, 23, 2505–2510.
[https://doi.org/10.3748/wjg.v23.i14.2505]

-
Zhu, P.; Li, J.; Fu, X.; Yu, Z. Phytomedicine 2019, 59, 152760.
[https://doi.org/10.1016/j.phymed.2018.11.020]

-
Yang, K.; Qiu, J.; Huang, Z.; Yu, Z.; Wang, W.; Hu, H.; You, Y. J. Ethnopharmacol. 2022, 284, 114759.
[https://doi.org/10.1016/j.jep.2021.114759]

- Zhang, M. X.; Huang, G. Y.; Bai, Y. Q.; Li, H.; Yang, B. Zhongguo Zhong Yao Za Zhi 2021, 46, 1017–1025.
-
Mai, N. T.; Doan, V. V.; Lan, H. T. T.; Anh, B. T. M.; Hoang, N. H.; Tai, B. H.; Nhiem, N. X.; Yen, P. H.; Park, S. J.; Seo, Y.; Namkung, W.; Kim, S. H.; Kiem, P. V. Nat. Prod. Res. 2021, 35, 3360–3369.
[https://doi.org/10.1080/14786419.2019.1700247]

-
Zhang, F.; Zhai, J.; Weng, N.; Gao, J.; Yin, J.; Chen, W. Front. Pharmacol. 2022, 13, 816036.
[https://doi.org/10.3389/fphar.2022.816036]

-
Huang, S.; Zhang, D.; Li, Y.; Fan, H.; Liu, Y.; Huang, W.; Deng, C.; Wang, W.; Song, X. Am. J. Chinese Med. 2021, 49, 1577–1622.
[https://doi.org/10.1142/S0192415X21500749]

-
Chen, X.; Tang, R.; Liu, T.; Dai, W.; Liu, Q.; Gong, G.; Song, S.; Hu, M.; Huang, L.; Wang, Z. Int. J. Biol. Macromol. 2019, 131, 744–751.
[https://doi.org/10.1016/j.ijbiomac.2019.03.129]

-
Fan, S.; Liu, C.; Jiang, Y.; Gao, Y.; Chen, Y.; Fu, K.; Yao, X.; Huang, M.; Bi, H. J. Ethnopharmacol. 2019, 245, 112103.
[https://doi.org/10.1016/j.jep.2019.112103]

-
Chen, Z. Liu, F.; Zheng, N.; Guo, M.; Bao, L.; Zhan, Y.; Zhang, M.; Zhao, Y.; Guo, W.; Ding, G. Biomed. Pharmacother. 2019, 110, 285–293.
[https://doi.org/10.1016/j.biopha.2018.11.069]

- Liu, J.; Lin H.; Yuan, L.; Wang, D.; Wang, C.; Sun, J.; Zhang, C.; Chen, J.; Li, H.; Jing, S. eCAM 2021, 2021, 9998982.
-
Teng, F.; Wang, W.; Zhang, W.; Qu, J.; Liu, B.; Chen, J.; Liu, S.; Li, M.; Chen, W.; Wei, H. Biopharm. Drug Dispos. 2022, 43, 119–129.
[https://doi.org/10.1002/bdd.2312]

-
Vo, P. H. T.; Nguyen, T. D. T.; Tran, H. T.; Nguyen, Y. N.; Doan, M. T.; Nguyen, P. H.; Lien, G. T. K.; To, D. C.; Tran, M. H. Bioorg. Med. Chem. Lett. 2021, 31, 127673.
[https://doi.org/10.1016/j.bmcl.2020.127673]

-
Jung, Y. W.; Lee, B. M.; Ha, M. T.; Tran, M. H.; Kim, J. A.; Lee, S.; Lee, J. H.; Woo, M. H.; Min, B. S. Arch. Pharm. Res. 2019, 42, 332–343.
[https://doi.org/10.1007/s12272-018-1093-4]

-
Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. Nucleic Acids Res. 2018, 46(W1), W363–W367.
[https://doi.org/10.1093/nar/gky473]

-
Lipinski, C. A. J. Pharmacol. Toxicol. Methods 2000, 44, 235–249.
[https://doi.org/10.1016/S1056-8719(00)00107-6]

-
Daina, A.; Michielin, O.; Zoete, V. Sci. Rep. 2017, 7, 42717.
[https://doi.org/10.1038/srep42717]

-
Mukherjee, A.; Su, A.; Rajan, K. J. Chem. Inf. Model. 2021, 61, 2187–2197.
[https://doi.org/10.1021/acs.jcim.0c01409]

-
Liu, K. T.; Lesca, P. Chem. Biol. Interact. 1982, 41, 39–47.
[https://doi.org/10.1016/0009-2797(82)90015-1]

-
Ikeya, Y.; Mitsuhashi, H.; Sasaki, H.; Matsuzaki, Y.; Matsuzaki, T.; Hosoya, E. Chem. Pharm. Bull. 1990, 38, 136–141.
[https://doi.org/10.1248/cpb.38.136]

-
Ikeya, Y.; Taguchi, H.; Yosioka, I.; Iitaka, Y.; Kobayashi, H. Chem. Pharm. Bull. 1979, 27, 1395–1401.
[https://doi.org/10.1248/cpb.27.1395]

-
Ikeya, Y.; Taguchi, H.; Yosioka, I.; Kobayashi, H. Chem. Pharm. Bull. 1979, 27, 1383–1394.
[https://doi.org/10.1248/cpb.27.1383]

-
Nhan, N. T.; Nguyen, P. H.; Tran, M. H.; Nguyen, P. D.; Tran, D. T.; To, D. C. J. Asian Nat. Prod. Res. 2021, 23, 414–422.
[https://doi.org/10.1080/10286020.2020.1767079]

-
Ferrer, M. D.; Busquets-Cortés, C.; Capó, X.; Tejada, S.; Tur, J. A.; Pons, A.; Sureda, A. Curr. Med. Chem. 2019, 26, 3225–3241.
[https://doi.org/10.2174/0929867325666180514112124]

-
Jagannathan, R. ACS Omega 2019, 4, 5402–5411.
[https://doi.org/10.1021/acsomega.8b01764]