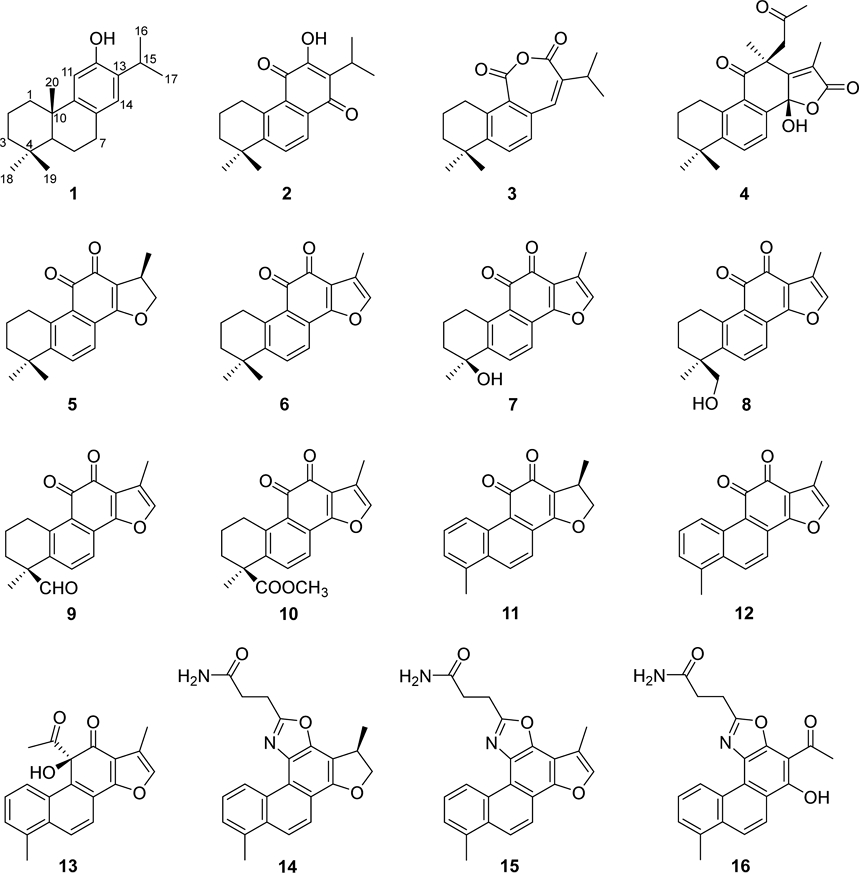
Inhibition of α-Glucosidase by Abietane-Type Diterpenoids Isolated from Roots of Salvia miltiorrhiza
Abstract
Salvia miltiorrhiza is a traditional medicinal plant used in Asian medicine for various therapeutic purposes. This plant contains numerous bioactive secondary metabolites, particularly abietane-type diterpenoids. In this study, 16 abietane-type diterpenoids were isolated from S. miltiorrhiza root extracts and structurally identified through advanced spectroscopic techniques. Among them, tanshinone IIA (6) and 15,16-dihydrotanshinone I (11) exhibited potent α-glucosidase inhibition, with IC50 values of 48.38 ± 0.57 and 48.02 ± 0.47 µM, respectively. Enzyme kinetic studies revealed that these compounds served as non-competitive inhibitors of α-glucosidase. Our findings indicate that natural compounds from S. miltiorrhiza show promise as safe and effective α-glucosidase inhibitors, providing an alternative approach to diabetes treatment. This study contributes to the growing interest in utilizing natural sources for α-glucosidase inhibition and their potential application in healthcare and disease management.
Keywords:
Salvia miltiorrhiza, diterpenoid, α-glucosidase inhibition, enzyme kineticsIntroduction
α-Glucosidase, a crucial enzyme in carbohydrate digestion, is primarily located at the border of the small intestine, where it acts on glycosidic bonds.1 This enzyme plays a key role in converting carbohydrates into absorbable monosaccharides.2,3 α-Glucosidase inhibitors are oral anti-diabetic medications used to manage type 2 diabetes by preventing carbohydrate digestion.4 Given its important role in metabolizing polysaccharides, particularly in regulating plasma blood levels, the investigation of α-glucosidase activity and its inhibition holds significant interest. Several α-glucosidase inhibitors, such as miglitol, voglibose, and acarbose, have been employed as antidiabetic drugs.5,6 These drugs function by inhibiting α-glucosidase activity, preventing the release of digested single sugars during carbohydrate digestion.7 However, these medications have been associated with side effects, such as stomach discomfort and diarrhea.8 Consequently, the discovery of potential α-glucosidase inhibitors with no side effects and safety for health, especially from natural sources, is highly necessary. Furthermore, α-glucosidase also participates in immune responses related to various diseases, including Pompe disease,9 P-glycoprotein trafficking,10 and cancer.11
Salvia miltiorrhiza Bunge, a perennial plant belonging to the Salvia genus in the Lamiaceae family, has a long history of use.12 Its roots and rhizomes have been employed in ethnopharmacology for over a thousand years and have been officially documented in the Chinese pharmacopeia since 1963.13 Traditional Asian medicine has reported various therapeutic properties of this plant, including enhancing blood circulation, reducing inner stress, and treating cardiovascular and cerebrovascular diseases as well as chronic renal failure.14-16 In addition, S. miltiorrhiza is frequently used with other herbs to treat various diseases, including diabetes,17 coronary heart disease,18 osteoporosis,19 Alzheimer’s,20 and cancer.21 Phytochemical analysis has revealed that the major components contained in this plant are lipophilic diterpenoids, including various tanshinone analogues, as well as hydrophilic phenolic compounds like salvianolic acids and their derivatives.12,22,23 Tanshinones isolated from S. miltiorrhiza, including cryptotanshinone, tanshinol B, and dehydrodanshenol A, were previously reported to exhibit significant protein tyrosine phosphatase 1B (PTP1B) inhibition, with IC50 values ranging from 4.7 to 8.5 μM.24 Furthermore, these active compounds could tightly bind to the allosteric site of the target enzyme through in silico studies.
In our ongoing efforts to identify bioactive secondary metabolites from Korean medicinal plants, we successfully isolated 16 compounds from the roots of S. miltiorrhiza. The structures of all isolated compounds were determined using state-of-the-art spectroscopic techniques. Moreover, the in vitro α-glucosidase inhibitory activity of all isolated compounds was evaluated. To gain a more comprehensive understanding of the inhibition mechanism and the inhibition constant, kinetic studies involving the active compounds and the α-glucosidase enzyme were conducted.
Experimental
General experimental procedures – Experiments in this study were conducted similarly to our previously described procedures.25-27 Compound isolation processes were carried out through column chromatography (CC) using silica gel 60 (0.063-0.200 mm) and RP-18 (40-63 μM) (Merck, Darmstadt, Germany). HPLC procedures were performed on a Waters Alliance HPLC system (MA, USA) featuring a 1525 Binary pump, a Waters 2998 PDA detector, and a C18 column (21.2 × 250 mm, 10 μM). NMR data were obtained on a Bruker Avance Digital 500 MHz spectrometer (Bruker, Karlsruhe, Germany), J in Hz. TLC was conducted using glass plates precoated with silica gel 60 F254 and RP-18 F254s (Merck). The TLC plates were examined under UV light at 254 nm and 356 nm to visualize the isolated compounds. Additionally, the plates were visualized by spraying them with a 10% sulfuric acid solution, followed by heating at 110oC for 1 min.
Plant material – S. miltiorrhiza roots were obtained from the Kyungdong market (Dongdaemun-gu, Seoul) in February 2015. The plant was authenticated by Professor Byung Sun Min, Daegu Catholic University, and deposited at the Pharmacognosy laboratory, College of Pharmacy, Kyungpook National University (voucher code: 18ASM6).
Extraction and isolation – The dried roots of S. miltiorrhiza (10.0 kg) were sliced into small pieces and extracted three times using MeOH (15 L × 3 h) under reflux conditions. After concentration, the MeOH residue (1.0 kg) was dissolved with 2L of water and subsequently partitioned with CHCl3 and EtOAc to obtain CHCl3 (74.3 g) and EtOAc (55.4 g) extracts, respectively. The detailed isolation procedure was described in our previous study.12
Ferruginol (1) – Amorphous powder; 1H-NMR (CDCl3, 500 MHz): δH 2.17 (1H, m, H-1a), 1.39 (1H, dd, J = 13.1, 3.6 Hz, H-1b), 1.85 (1H, m, H-2a), 1.68 (1H, m, H-2b), 1.47 (1H, m, H-3a), 1.20 (1H, m, H-3b), 1.32 (1H, m, H-5), 1.73 (1H, m, H-6a), 1.63 (1H, m, H-6b), 2.82 (2H, m, H-7), 6.63 (1H, br s, H-11), 6.83 (1H, s, H-14), 3.11 (1H, m, H-15), 1.23 (each 3H, dd, J = 8.2, 6.9 Hz, H-16, H-17), 0.91 (3H, s, H-18), 0.94 (3H, s, H-19), 1.17 (3H, d, J = 0.4 Hz, H-20); 13C-NMR (CDCl3, 125 MHz): δC 39.0 (C-1), 19.4 (C-2), 41.8 (C-3), 33.6 (C-4), 50.5 (C-5), 19.5 (C-6), 29.9 (C-7), 127.5 (C-8), 148.8 (C-9), 37.7 (C-10), 111.1 (C-11), 150.8 (C-12), 131.5 (C-13), 126.8 (C-14), 27.0 (C-15), 22.9 (C-16), 22.7 (C-17), 21.8 (C-18), 33.5 (C-19), 24.9 (C-20).28
Deoxyneocryptotanshinone (2) – Amorphous powder; 1H-NMR (CDCl3, 500 MHz): δH 3.24 (2H, t, J = 6.4 Hz, H-1), 1.83 (2H, m, H-2), 1.69 (2H, m, H-3), 7.88 (1H, d, J = 8.2 Hz, H-6), 7.94 (1H, d, J = 8.2 Hz, H-7), 3.37 (1H, m, H-15), 1.25 (3H, s, H-16), 1.27 (3H, s, H-17), 1.33 (each 3H, s, H-18, H-19); 13C-NMR (CDCl3, 125 MHz): δC 29.7 (C-1), 19.0 (C-2), 37.7 (C-3), 34.6 (C-4), 152.2 (C-5), 133.1 (C-6), 125.4 (C-7), 132.6 (C-8), 126.9 (C-9), 139.9 (C-10), 183.2 (C-11), 154.4 (C-12), 124.3 (C-13), 184.1 (C-14), 24.2 (C-15), 19.3 (C-16), 19.3 (C-17), 31.1 (C-18), 31.1 (C-19).29
Normiltirone (3) – Amorphous powder; 1H and 13C NMR data, see reference.12
Salviprzol A (4) – Amorphous powder; 1H-NMR (CDCl3, 500 MHz): δH 3.23 (1H, m, H-1a), 2.97 (1H, m, H-1b), 1.90 (1H, m, H-2a), 1.73 (1H, m, H-2b), 1.74 (2H, m, H-3), 7.80 (1H, d, J = 8.3 Hz, H-6), 7.77 (1H, d, J = 8.3 Hz, H-7), 2.06 (3H, s, H-17), 3.28 (1H, d, J = 13.6 Hz, H-18a), 2.92 (1H, d, J = 13.6 Hz, H-18b), 2.40 (3H, s, H-20), 1.33 (each 3H, s, H-21, H-22); 13C-NMR (CDCl3, 125 MHz): δC 29.9 (C-1), 19.4 (C-2), 38.0 (C-3), 34.6 (C-4), 149.7 (C-5), 134.6 (C-6), 124.2 (C-7), 138.3 (C-8), 125.1 (C-9), 140.7 (C-10), 195.9 (C-11), 80.6 (C-12), 152.9 (C-13), 99.8 (C-14), 128.1 (C-15), 170.5 (C-16), 9.3 (C-17), 49.5 (C-18), 209.7 (C-19), 33.5 (C-20), 32.0 (C-21), 32.0 (C-22).30
Cryptotanshinone (5) – Amorphous powder; 1H-NMR (CDCl3, 500 MHz): δH 3.22 (2H, t, J = 6.4 Hz, H-1), 1.79 (2H, m, H-2), 1.65 (2H, m, H-3), 7.63 (1H, d, J = 8.2 Hz, H-6), 7.49 (1H, d, J = 8.2 Hz, H-7), 4.88 (1H, t, J = 9.5 Hz, H-15a), 4.36 (1H, dd, J = 9.5, 6.0 Hz, H-15b), 3.59 (1H, m, H-16), 1.35 (3H, d, J = 6.8 Hz, H-17), 1.30 (each 3H, s, H-18, H-19); 13C-NMR (CDCl3, 125 MHz): δC 29.8 (C-1), 19.2 (C-2), 38.0 (C-3), 34.9 (C-4), 143.9 (C-5), 132.7 (C-6), 122.7 (C-7), 128.5 (C-8), 126.5 (C-9), 152.4 (C-10), 184.4 (C-11), 175.9 (C-12), 118.4 (C-13), 171.1 (C-14), 81.6 (C-15), 34.7 (C-16), 19.0 (C-17), 32.1 (C-18), 32.2 (C-19).31
Tanshinone IIA (6) – Amorphous powder; 1H-NMR (CDCl3, 500 MHz): δH 3.18 (2H, t, J = 6.4 Hz, H-1), 1.81 (2H, m, H-2), 1.63 (2H, m, H-3), 7.63 (1H, d, J = 8.2 Hz, H-6), 7.55 (1H, d, J = 8.2 Hz, H-7), 7.22 (1H, q, J = 1.2 Hz, H-15), 2.26 (3H, d, J = 1.3 Hz, H-17), 1.31 (each 3H, s, H-18, H-19); 13C-NMR (CDCl3, 125 MHz): δC 29.9 (C-1), 19.2 (C-2), 38.0 (C-3), 34.9 (C-4), 144.4 (C-5), 133.5 (C-6), 120.4 (C-7), 127.7 (C-8), 126.5 (C-9), 150.4 (C-10), 183.9 (C-11), 175.9 (C-12), 120.0 (C-13), 161.9 (C-14), 141.4 (C-15), 121.3 (C-16), 8.9 (C-17), 32.1 (C-18), 32.1 (C-19).31
Tanshinol B (7) – Amorphous powder; 1H-NMR (CDCl3, 500 MHz): δH 3.10 (2H, m, H-1), 1.67 (1H, m, H-2a), 1.87 (1H, m, H-2b), 1.80 (2H, m, H-3), 7.95 (1H, d, J = 7.9 Hz, H-6), 7.60 (1H, d, J = 7.9 Hz, H-7), 7.71 (1H, br s, H-15), 2.16 (3H, s, H-17), 1.40 (3H, s, H-18); 13C-NMR (CDCl3, 125 MHz): δC 28.7 (C-1), 19.5 (C-2), 37.6 (C-3), 69.0 (C-4), 148.3 (C-5), 133.5 (C-6), 119.9 (C-7), 127.6 (C-8), 125.8 (C-9), 142.4 (C-10), 182.5 (C-11), 175.0 (C-12), 120.1 (C-13), 160.4 (C-14), 142.3 (C-15), 119.6 (C-16), 8.5 (C-17), 31.3 (C-18).32
Tanshinone IIB (8) – Amorphous powder; 1H-NMR (CDCl3, 500 MHz): δH 3.17 (2H, dt, J = 13.5, 6.5 Hz, H-1), 7.65 (1H, d, J = 8.1 Hz, H-6), 7.52 (1H, d, J = 8.1 Hz, H-7), 7.21 (1H, br s, H-15), 2.26 (3H, s, H-17), 1.27 (3H, s, H-18), 3.79 (1H, d, J = 11.1 Hz, H-19a), 3.61 (1H, d, J = 11.1 Hz, H-19b); 13C-NMR (CDCl3, 125 MHz): δC 29.8 (C-1), 19.0 (C-2), 32.4 (C-3), 40.3 (C-4), 146.3 (C-5), 133.8 (C-6), 119.9 (C-7), 127.8 (C-8), 126.7 (C-9), 146.1 (C-10), 183.4 (C-11), 175.6 (C-12), 121.3 (C-13), 161.6 (C-14), 120.4 (C-15), 141.4 (C-16), 8.9 (C-17), 26.9 (C-18), 71.6 (C-19).33
Tanshinonal (9) – Amorphous powder; 1H-NMR (CDCl3, 500 MHz): δH 3.24 (2H, m, H-1), 1.85 (2H, dd, J = 12.3, 6.3 Hz, H-2), 2.07 (1H, m, H-3a), 1.66 (1H, m, H-3b), 7.33 (1H, d, J = 8.1 Hz, H-6), 7.60 (1H, d, J = 8.1 Hz, H-7), 7.25 (1H, d, J = 1.1 Hz, H-15), 2.27 (3H, d, J = 0.9 Hz, H-17), 1.46 (3H, s, H-18), 9.49 (1H, br s, H-19); 13C-NMR (CDCl3, 125 MHz): δC 29.2 (C-1), 18.7 (C-2), 30.1 (C-3), 51.3 (C-4), 139.9 (C-5), 135.5 (C-6), 120.7 (C-7), 129.0 (C-8), 127.0 (C-9), 145.6 (C-10), 183.4 (C-11), 175.5 (C-12), 121.5 (C-13), 161.1 (C-14), 120.7 (C-15), 141.9 (C-16), 8.9 (C-17), 24.1 (C-18), 201.8 (C-19).34
Methyl tanshinonate (10) – Amorphous powder; 1H-NMR (CDCl3, 500 MHz): δH 3.23 (2H, m, H-1), 2.29 (1H, m, H-2a), 1.74 (1H, m, H-2b), 1.82 (2H, td, J = 13.7, 6.8 Hz, H-3), 7.56 (1H, d, J = 8.1 Hz, H-6), 7.48 (1H, d, J = 8.1 Hz, H-7), 7.23 (1H, br s, H-15), 2.26 (3H, s, H-17), 1.58 (3H, s, H-18), 3.67 (3H, s, H-20); 13C-NMR (CDCl3, 125 MHz): δC 29.2 (C-1), 19.3 (C-2), 34.2 (C-3), 47.3 (C-4), 143.2 (C-5), 135.1 (C-6), 120.4 (C-7), 126.7 (C-8), 128.7 (C-9), 141.7 (C-10), 183.5 (C-11), 175.6 (C-12), 121.4 (C-13), 161.5 (C-14), 120.4 (C-15), 144.5 (C-16), 8.9 (C-17), 27.7 (C-18), 177.1 (C-19), 52.6 (C-20).34
15,16-Dihydrotanshinone I (11) – Amorphous powder; 1H-NMR (CDCl3, 500 MHz): δH 9.28 (1H, d, J = 8.9 Hz, H-1), 7.56 (1H, dd, J = 8.9, 6.9 Hz, H-2), 7.39 (1H, d, J = 6.9 Hz, H-3), 8.29 (1H, dd, J = 8.7, 0.9 Hz, H-6), 7.74 (1H, d, J = 8.7 Hz, H-7), 4.97 (1H, t, J = 9.5 Hz, H-15a), 4.43 (1H, dd, J = 9.5, 6.3 Hz, H-15b), 3.65 (1H, m, H-16), 1.41 (3H, d, J = 6.8 Hz, H-17), 2.69 (3H, s, H-18); 13C-NMR (CDCl3, 125 MHz): δC 126.3 (C-1), 130.6 (C-2), 128.4 (C-3), 135.1 (C-4), 134.8 (C-5), 132.2 (C-6), 118.8 (C-7), 129.0 (C-8), 125.5 (C-9), 132.1 (C-10), 184.4 (C-11), 175.6 (C-12), 120.4 (C-13), 170.7 (C-14), 81.8 (C-15), 34.9 (C-16), 18.9 (C-17), 20.0 (C-18).35
Tanshinone I (12) – Amorphous powder; 1H-NMR (CDCl3, 500 MHz): δH 9.24 (1H, d, J = 8.9 Hz, H-1), 7.54 (1H, dd, J = 8.9, 6.9 Hz, H-2), 7.29 (1H, d, J = 6.9 Hz, H-3), 8.28 (1H, d, J = 8.7 Hz, H-6), 7.79 (1H, d, J = 8.7 Hz, H-7), 7.34 (1H, d, J = 6.9 Hz, H-15), 2.28 (3H, d, J = 1.3 Hz, H-17), 2.68 (3H, s, H-18); 13C-NMR (CDCl3, 125 MHz): δC 124.9 (C-1), 130.8 (C-2), 128.5 (C-3), 135.1 (C-4), 134.1 (C-5), 132.9 (C-6), 118.8 (C-7), 129.8 (C-8), 123.5 (C-9), 132.9 (C-10), 183.6 (C-11), 175.8 (C-12), 121.9 (C-13), 161.3 (C-14), 142.2 (C-15), 120.6 (C-16), 8.9 (C-17), 20.0 (C-18).35
Dehydrodanshenol A (13) – Amorphous powder; 1H-NMR (CDCl3, 500 MHz): δH 9.02 (1H, d, J = 8.8 Hz, H-1), 7.50 (1H, m, H-2), 7.40 (1H, d, J = 6.9 Hz, H-3), 8.12 (1H, d, J = 8.8 Hz, H-6), 7.93 (1H, d, J = 8.8 Hz, H-7), 7.34 (1H, br s, H-15), 2.33 (3H, s, H-17), 2.73 (3H, s, H-18), 3.36 (1H, d, J = 13.5 Hz, H-19a), 3.05 (1H, d, J = 13.5 Hz, H-19b), 2.03 (3H, s, H-21); 13C-NMR (CDCl3, 125 MHz): δC 125.4 (C-1), 127.0 (C-2), 128.0 (C-3), 135.4 (C-4), 134.1 (C-5), 126.1 (C-6), 118.8 (C-7), 120.8 (C-8), 137.1 (C-9), 131.5 (C-10), 80.7 (C-11), 197.4 (C-12), 120.3 (C-13), 162.4 (C-14), 116.7 (C-15), 141.6 (C-16), 8.9 (C-17), 20.4 (C-18), 56.3 (C-19), 205.3 (C-20), 32.2 (C-21).36
Isosalviamide F (14) – Amorphous powder; 1H and 13C NMR data, see reference.12
Isosalviamide G (15) – Amorphous powder; 1H and 13C NMR data, see reference.12
Isosalviamide H (16) – Amorphous powder; 1H and 13C NMR data, see reference.12
Inhibitory activity on α-glucosidase – The assay for inhibiting α-glucosidase was carried out following established procedures described in previous studies.5,37,38 To initiate the enzymatic reaction, 20 μL of α-glucosidase enzyme from Saccharomyces cerevisiae (EC 3.2.1.20, G5003; Sigma-Aldrich, MO, USA) at a concentration of 0.1 unit/mL was added into a 96-well plate, which contained 20 μL of test compounds and 20 μL of phosphate buffer (100 mM, pH = 6.8). Afterward, the enzyme reaction was started by adding 20 μL substrate p-NPG (2.5 mM) and incubated at 37oC. After 10 min, 80 μL Na2CO3 (0.2 M) was used for each well to finalize the reaction. The absorbance was measured at 405 nm using a PHOmo microplate reader (Autobio Labtec Instruments, Zhengzhou, China). Acarbose (A8980; Sigma-Aldrich) was used as a positive control. The percentage of α-glucosidase inhibitory activity was calculated using the following equation:
Inhibitory activity (%) = [(ΔControl − ΔInhibitor)/ΔControl] × 100, where ΔControl and ΔInhibitor represent the signal intensities of the control and inhibitor solutions after a 10 min incubation period, respectively.
The determination and calculation of IC50 values were performed using Microsoft Excel (Microsoft 365, WA, USA). The results were expressed as the means ± standard error based on data from at least three experiments. Statistical significance (p < 0.05) was calculated through the application of Duncan’s tests and ANOVA.
Enzyme kinetics – To understand the inhibition mechanism of α-glucosidase by active compounds, two graphical kinetic methods, including Lineweaver–Burk and Dixon plots, were carried out.39 The Dixon plot was employed to assess enzymatic reactions across different concentrations of substrate, specifically 0.625, 1.25, and 2.5 mM. The inhibition constant (Ki) was determined via the Dixon plot, where the x-axis value was taken as -Ki.
In addition, the Lineweaver–Burk plot was employed to identify the α-glucosidase inhibition mode of test compounds through various concentrations (0, 19.75, 48, and 83.75 μM for compound 6; and 0, 25.25, 55.25, and 100 μM for compound 11).
Results and Discussion
S. miltiorrhiza roots were extracted with methanol. After removing the solvent, the residue was mixed with water and partitioned into CHCl3 and EtOAc extracts. These fractions were separated using chromatography techniques, leading to the isolation of 16 compounds (Fig. 1). Their chemical structures were elucidated through NMR spectroscopic analysis, including 1H and 13C-NMR, as well as literature comparisons: ferruginol (1),28 deoxyneocryptotanshinone (2),29 normiltirone (3),12 salviprzol A (4),30 cryptotanshinone (5),31 tanshinone IIA (6),31 tanshinol B (7),32 tanshinone IIB (8),33 tanshinonal (9),34 methyl tanshinonate (10),34 15,16-dihydrotanshinone I (11),35 tanshinone I (12),35 dehydrodanshenol A (13),36 and isosalviamides F–H (14–16).12
Compound 6 was yielded as an amorphous powder. The 1H-NMR spectrum of 6 displayed signals characteristic of an abietane-type diterpenoid, including three methyl groups at δH 2.26 (3H, d, J = 1.3 Hz, H-17) and 1.31 (each 3H, s, H-18, H-19), three methylene groups at δH 3.18 (2H, t, J = 6.4 Hz, H-1), 1.81 (2H, m, H-2), and 1.63 (2H, m, H-3), a methine proton at δH 7.22 (1H, q, J = 1.2 Hz, H-15), and two aromatic protons at δH 7.63 (1H, d, J = 8.2 Hz, H-6) and 7.55 (1H, d, J = 8.2 Hz, H-7). In addition, the 13C-NMR and DEPT spectra of compound 6 exhibited signals of two carbonyl groups at δC 183.9 (C-11) and 175.9 (C-12), nine olefinic carbons at δC 161.9, 150.4, 144.4, 141.4, 133.5, 127.7, 126.5, 121.3, 120.4, and 120.0, and three methyl groups at δC 32.1, 32.1 and 8.9. The position of the methyl group at C-17 was determined based on the HMBC correlations of H-17 with C-13, C-15, and C-16, whereas the positions of the two remaining methyl groups were determined based on the HMBC correlations of H-18 and H-19 with C-3, C-4, and C-5. This analysis, along with published data from the literature, led to the elucidation of compound 6 as tanshinone IIA.31
Compound 11 was obtained as an amorphous powder. The 1H-NMR spectrum of this compound showed the presence of two methyl groups at δH 1.41 (3H, d, J = 6.8 Hz, H-17) and 2.69 (3H, s, H-18), a methylene group at δH 4.97 (1H, t, J = 9.5 Hz, H-15a) and 4.43 (1H, dd, J = 9.5, 6.3 Hz, H-15b), and five aromatic protons at δH 9.28 (1H, d, J = 8.9 Hz, H-1), 7.56 (1H, dd, J = 8.9, 6.9 Hz, H-2), 7.39 (1H, d, J = 6.9 Hz, H-3), 8.29 (1H, dd, J = 8.7, 0.9 Hz, H-6), and 7.74 (1H, d, J = 8.7 Hz, H-7). They correlated with the carbons resonating at δH 18.9, 20.0, 81.8, 126.3, 130.6, 128.4, 132.2, and 118.8 in the HMQC spectrum, respectively. Furthermore, the 13C-NMR spectrum of compound 11 showed signals of two carbonyl groups at δC 184.4 (C-11) and 175.6 (C-12), and seven quaternary carbons at δC 135.1 (C-4), 134.8 (C-5), 129.0 (C-8), 125.5 (C-9), 132.1 (C-10), 120.4 (C-13), and 170.7 (C-14). This evidence, similar to that reported for compound 6, indicates that compound 11 is an abietane-type diterpenoid. Compared with the literature, compound 11 was clearly identified as 15,16-dihydrotanshinone I.35
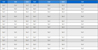
The examination of their ability to inhibit α-glucosidase activity was evaluated on all isolated compounds (1–16). For reference, acarbose, a well-known α-glucosidase inhibitor, served as positive control and exhibited an IC50 value of 222.62 ± 1.98 μM (Table 1).
Compounds 2 and 3 demonstrated moderate α-glucosidase inhibition, with IC50 values of 58.06 ± 0.97 and 67.06 ± 0.97 μM, respectively. Compound 6, which had an additional double bond at C-15 and C-16 in comparison to compound 5, displayed α-glucosidase inhibitory activity with an IC50 value of 48.38 ± 0.57 μM, while compound 5 showed no α-glucosidase inhibition. This suggests that the double bond at C-15 and C-16 may contribute to the inhibitory activity of abietane diterpenoids against α-glucosidase.
Compounds 7–10 shared a similar structure with compound 6, except for the differences in the functional groups at C-19. They exhibited weaker α-glucosidase inhibitory activity compared to compound 6, with IC50 values ranging from 55.13 to more than 100 μM. This evidence considers the idea that less polar substituents in the structure of abietane diterpenoids may influence α-glucosidase inhibition.
Compounds 11 and 12, which had an aromatic ring in ring A and lacked a methyl group linked to C-4, exhibited α-glucosidase inhibition with IC50 values of 48.02± 0.47 and 51.69 ± 0.86 μM, respectively. The remaining compounds (13–16) showed no α-glucosidase inhibitory effect.
In our research framework, we have chosen the two most potent active compounds (6 and 11) to continue investigating the inhibition mechanism and determining the inhibition constant (Ki) through enzyme kinetics studies. Lineweaver–Burk and Dixon plots were employed. The Lineweaver–Burk graph showed that the spot where the lines of the inhibitors crossed on the x-axis or y-axis indicated whether the inhibition was non-competitive or competitive respectively.39,40
As presented in Fig. 2, compounds 6 and 11 exhibited a family of intersecting straight lines on the x-axis (1/[S]-axis), indicating that these active compounds served as a non-competitive inhibitor of α-glucosidase. The Ki values for compounds 6 and 11 were determined through Dixon plots and calculated to be 47.17 ± 0.41 and 45.41 ± 0.40 μM, respectively.
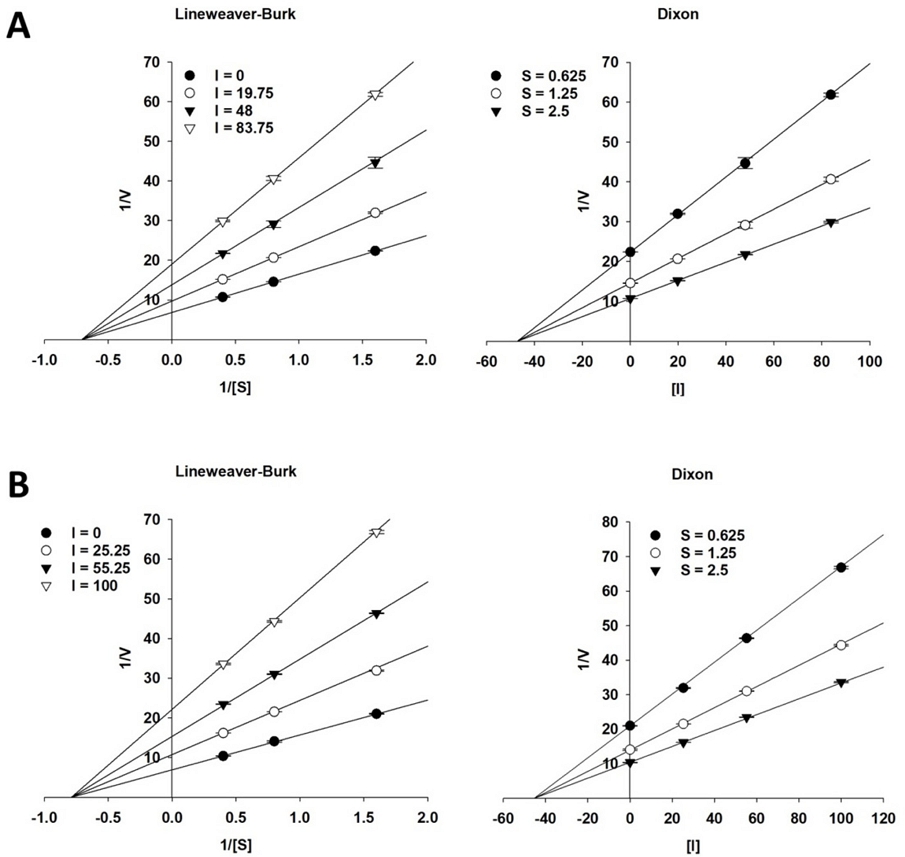
Lineweaver−Burk and Dixon plots for the inhibition of α-glucosidase by active compounds 6 (A) and 11 (B).
In conclusion, 16 abietane-type diterpenoids (1–16) were isolated from extracts of S. miltiorrhiza roots. The structures of all compounds were determined through extensive spectroscopic analysis, including 1H- and 13C-NMR, in conjunction with data from existing literature. Our investigation into α-glucosidase inhibitory activity revealed that tanshinone IIA (6) and 15,16-dihydrotanshinone I (11) exhibited the most potent α-glucosidase inhibition, with IC50 values of 48.38 ± 0.57 and 48.02±0.47 μM, respectively, surpassing the positive control acarbose (IC50 = 222.62 ± 1.98 μM). Our kinetic studies demonstrated that these active compounds acted as non-competitive inhibitors of α-glucosidase. These promising results establish that natural compounds from S. miltiorrhiza with α-glucosidase inhibition potential could serve as a safe and primary complement to treating diabetes and related diseases. These leading secondary metabolites may warrant further exploration through in silico approaches or in vivo investigations of their α-glucosidase inhibitory activities using animal models.
Acknowledgments
This research was supported by the National Research Foundation of Korea (NRF) funded by the Korea government (MSIT) (No. NRF-2020R1A5A2017323). The authors would like to thank Korea Basic Science Institute (KBSI) Daegu Center for the mass spectra measurement service.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
-
Wan, J.-X.; Lim, G.; Lee, J.; Sun, X.-B.; Gao, D.-Y.; Si, Y.-X.; Shi, X.-L.; Qian, G.-Y.; Wang, Q.; Park, Y.-D. Int. J. Biol. Macromol. 2019, 124, 771-779.
[https://doi.org/10.1016/j.ijbiomac.2018.11.268]

-
Mohamed Sham Shihabudeen, H.; Hansi Priscilla, D.; Thirumurugan, K. Nutr. Metab. 2011, 8, 46.
[https://doi.org/10.1186/1743-7075-8-46]

-
Le, T. T.; Ha, M. T.; Hoang, L. M.; Vu, N. K.; Kim, J. A.; Min, B. S. Nat. Prod. Sci. 2022, 28, 143-152.
[https://doi.org/10.20307/nps.2022.28.3.143]

-
Ghauri, S.; Raza, S. Q.; Imran, M.; Saeed, S.; Rashid, M.; Naseer, R. 3 Biotech 2021, 11, 167.
[https://doi.org/10.1007/s13205-021-02717-8]

-
Phong, N. V.; Yang, S. Y.; Min, B. S.; Kim, J. A. J. Mol. Struct. 2023, 1282, 135188.
[https://doi.org/10.1016/j.molstruc.2023.135188]

-
Mushtaq, A.; Azam, U.; Mehreen, S.; Naseer, M. M. Eur. J. Med. Chem. 2023, 249, 115119.
[https://doi.org/10.1016/j.ejmech.2023.115119]

-
Siano, F.; Mamone, G.; Vasca, E.; Puppo, M. C.; Picariello, G. Food Res. Int. 2023, 170, 112962.
[https://doi.org/10.1016/j.foodres.2023.112962]

-
Yang, Y.; Yang, M.; Zhou, X.; Chen, H. Foods 2022, 11, 3530.
[https://doi.org/10.3390/foods11213530]

-
Li, H.-Y.; Lee, N.-C.; Chiu, Y.-T.; Chang, Y.-W.; Lin, C.-C.; Chou, C.-L.; Chien, Y.-H.; Hwu, W.-L.; Cheng, W.-C. Bioorg. Med. Chem. 2023, 78, 117129.
[https://doi.org/10.1016/j.bmc.2022.117129]

-
Barker, M. K.; Rose, D. R. J. Biol. Chem. 2013, 288, 13563-13574.
[https://doi.org/10.1074/jbc.M113.460436]

-
Smith, D. L.; Orlandella, R. M.; Allison, D. B.; Norian, L. A. GeroScience 2020, 43, 1123-1133.
[https://doi.org/10.1007/s11357-020-00278-x]

-
Ngo, T. M.; Tran, P. T.; Hoang, L. S.; Lee, J.-H.; Min, B. S.; Kim, J. A. Nat. Prod. Res. 2019, 35, 726-732.
[https://doi.org/10.1080/14786419.2019.1596098]

-
Jia, Q.; Zhu, R.; Tian, Y.; Chen, B.; Li, R.; Li, L.; Wang, L.; Che, Y.; Zhao, D.; Mo, F.; Gao, S.; Zhang, D. Phytomedicine 2019, 58, 152871.
[https://doi.org/10.1016/j.phymed.2019.152871]

-
Wang, L.; Ma, R.; Liu, C.; Liu, H.; Zhu, R.; Guo, S.; Tang, M.; Li, Y.; Niu, J.; Fu, M.; Gao, S.; Zhang, D. Curr. Pharm. Des. 2017, 23, 1077-1097.
[https://doi.org/10.2174/1381612822666161010105242]

-
Su, C.-Y.; Ming, Q.-L.; Rahman, K.; Han, T.; Qin, L.-P. Chin. J. Nat. Med. 2015, 13, 163-182.
[https://doi.org/10.1016/S1875-5364(15)30002-9]

-
Jung, I.; Kim, H.; Moon, S.; Lee, H.; Kim, B. Antioxidants 2020, 9, 857.
[https://doi.org/10.3390/antiox9090857]

-
Orgah, J. O.; He, S.; Wang, Y.; Jiang, M.; Wang, Y.; Orgah, E. A.; Duan, Y.; Zhao, B.; Zhang, B.; Han, J.; Zhu, Y. Pharmacol. Res. 2020, 153, 104654.
[https://doi.org/10.1016/j.phrs.2020.104654]

-
Ren, J.; Fu, L.; Nile, S. H.; Zhang, J.; Kai, G. Front. Pharmacol. 2019, 10, 753.
[https://doi.org/10.3389/fphar.2019.00753]

- Kim, D.-S.; Han, S.-H.; Lee, Y. C.; Park, S. J.; Yoo, W. H.; Kim, H.-R.; Chae, H.-J. Korean J. Clin. Pharm. 2009, 19, 75-80.
-
Park, B.; Song, H. S.; Kwon, J. E.; Cho, S. M.; Jang, S.-A.; Kim, M. Y.; Kang, S. C. BMC Complement. Altern. Med. 2017, 17, 545.
[https://doi.org/10.1186/s12906-017-2047-y]

-
Sung, B.; Chung, H. S.; Kim, M.; Kang, Y. J.; Kim, D. H.; Hwang, S. Y.; Kim, M. J.; Kim, C. M.; Chung, H. Y.; Kim, N. D. Exp. Ther. Med. 2015, 9, 1421-1428.
[https://doi.org/10.3892/etm.2015.2252]

-
Tang, H.; Liu, Y.; Yu, Z.; Sun, M.; Lin, L.; Liu, W.; Han, Q.; Wei, M.; Jin, Y. Front. Pharmacol. 2019, 10, 3389.
[https://doi.org/10.3389/fphar.2019.00373]

-
Jiang, Z.; Gao, W.; Huang, L. Front. Pharmacol. 2019, 10, 202.
[https://doi.org/10.3389/fphar.2019.00202]

-
Kim, D. H.; Paudel, P.; Yu, T.; Ngo, T. M.; Kim, J. A.; Jung, H. A.; Yokozawa, T.; Choi, J. S. Chem. Biol. Interact. 2017, 278, 65-73.
[https://doi.org/10.1016/j.cbi.2017.10.013]

-
Phong, N. V.; Chae, H. Y.; Oanh, V. T.; Min, B. S.; Kwon, M. J.; Kim, J. A. Nat. Prod. Sci. 2023, 29, 171-181.
[https://doi.org/10.20307/nps.2023.29.3.171]

-
Phong, N. V.; Oanh, V. T.; Yang, S. Y.; Choi, J. S.; Min, B. S.; Kim, J. A. Int. J. Biol. Macromol. 2021, 188, 719-728.
[https://doi.org/10.1016/j.ijbiomac.2021.08.091]

-
Oanh, V. T.; Phong, N. V.; Min, B. S.; Yang, S. Y.; Kim, J. A. J. Enzyme Inhib. Med. Chem. 2023, 38, 2251099.
[https://doi.org/10.1080/14756366.2023.2251099]

-
Ryu, Y. B.; Jeong, H. J.; Kim, J. H.; Kim, Y. M.; Park, J.-Y.; Kim, D.; Naguyen, T. T. H.; Park, S.-J.; Chang, J. S.; Park, K. H.; Rho, M.-C.; Lee, W. S. Bioorg. Med. Chem. 2010, 18, 7940-7947.
[https://doi.org/10.1016/j.bmc.2010.09.035]

-
Eghtesadi, F.; Moridi Farimani, M.; Hazeri, N.; Valizadeh, J. SpringerPlus 2016, 5, 1068.
[https://doi.org/10.1186/s40064-016-2652-0]

-
Xue, Y.; Wu, Y.; Zhu, H.; Li, X.-N.; Qian, J.-F.; Lai, Y.; Chen, C.; Yao, G.; Luo, Z.; Li, Y.; Zhang, Y. Fitoterapia 2014, 99, 204-210.
[https://doi.org/10.1016/j.fitote.2014.09.022]

-
Fronza, M.; Murillo, R.; Ślusarczyk, S.; Adams, M.; Hamburger, M.; Heinzmann, B.; Laufer, S.; Merfort, I. Bioorg. Med. Chem. 2011, 19, 4876-4881.
[https://doi.org/10.1016/j.bmc.2011.06.067]

-
Wang, F.; Yang, H.; Yu, S.; Xue, Y.; Fan, Z.; Liang, G.; Geng, M.; Zhang, A.; Ding, C. Org. Biomol. Chem. 2018, 16, 3376-3381.
[https://doi.org/10.1039/C8OB00567B]

-
Liu, A.-H.; Lin, Y.-H.; Yang, M.; Guo, H.; Guan, S.-H.; Sun, J.-H.; Guo, D.-A. J. Chromatogr. B 2007, 846, 32-41.
[https://doi.org/10.1016/j.jchromb.2006.08.002]

-
Lee, J.; Snyder, J. K. J. Org. Chem. 1990, 55, 4995-5008.
[https://doi.org/10.1021/jo00304a008]

-
Lee, S.-Y.; Choi, D.-Y.; Woo, E.-R. Arch. Pharm. Res. 2005, 28, 909-913.
[https://doi.org/10.1007/BF02973876]

-
Jiang, H.-L.; Wang, X.-Z.; Xiao, J.; Luo, X.-H.; Yao, X.-J.; Zhao, Y.-Y.; Chen, Y.-J.; Crews, P.; Wu, Q.-X. Tetrahedron 2013, 69, 6687-6692.
[https://doi.org/10.1016/j.tet.2013.05.115]

-
Phong, N. V.; Gao, D.; Kim, J. A.; Yang, S. Y. Metabolites 2023, 13, 557.
[https://doi.org/10.3390/metabo13040557]

-
Bongmo, L. V. L.; Bissoue, A. N.; Bissim, S. M.; Tabekoueng, G. B.; Tsopgni, W. D. T.; Lateef, M.; Kasali, F. M.; Ali, M. S.; Waffo, A. F. K.; Wansi, J. D. Nat. Prod. Sci. 2023, 29, 50-58.
[https://doi.org/10.20307/nps.2023.29.1.50]

-
Rodriguez, J.-M. G.; Hux, N. P.; Philips, S. J.; Towns, M. H. J. Chem. Educ. 2019, 96, 1833-1845.
[https://doi.org/10.1021/acs.jchemed.9b00396]

-
Park, S.-R.; Kim, Y. H.; Yang, S. Y. Nat. Prod. Sci. 2023, 29, 182-192.
[https://doi.org/10.20307/nps.2023.29.3.182]