Identification of Phenolic Compounds from Vietnamese Artichoke (Cynara scolymus L.) Leaf and Their Antioxidant Activities
Abstract
Artichoke (Cynara scolymus L.) is a perennial plant belonging to the Asteraceae family originating from the Mediterranean region. In Vietnam, there are some varieties of artichoke which are extensively cultivated and propagated in highland areas, however, there have been limited detailed scientific publications on the chemical composition and biological activity of artichoke grown in Vietnam. Therefore, this study provides a detailed description of the extraction, isolation, and structural determination of 20 natural secondary metabolites present in harvested artichoke. The antioxidant activity of the extract and the 9 isolated compounds are tested in 1,1-diphenyl-2-picrylhydrazyl radical scavenging and ex vivo malondialdehyde model. Among the selected compounds, 1-caffeoylquinic acid, 3-caffeoylquinic acid, chlorogenic acid, 4-caffeoylquinic acid, cynarin, 1,5-di-caffeoylquinic acid, 4,5-di-caffeoylquinic acid, cynaroside, and scolymoside exhibited strong radical scavenging activity with IC50 values ranging from 5.7 to 61.6 μM. In the malondialdehyde assay, 1,3-di-caffeoylquinic acid (or cynarin) showed the strongest activity with an IC50 value of 24.7 μM, followed by 1,5-di-caffeoylquinic acid (66.8 μM), and 4,5-di-caffeoylquinic acid (127.3 μM). This outcome contributes to establishing a database on the phytochemical and antioxidant activity of the Vietnamese artichoke.
Keywords:
Cynara scolymus L, Asteraceae, Vietnamese artichoke, Phenolic compound, AntioxidantIntroduction
Artichoke (Cynara scolymus L.) (also known as Cynara cardunculus L.) is a perennial plant belonging to the Asteraceae family originating from the Mediterranean region. Currently, artichoke is widely cultivated in many places around the world.1,2 In Vietnam, up until now, two varieties of artichoke are commonly grown and propagated in Da Lat, namely Artichoke A80 (“purple variety”, designated as AT) and Artichoke A85 (“green variety”, designated as AX).3 Traditionally, artichoke plants are propagated mainly by separating lateral shoots or using the same root from the previous crop.4 However, these methods can easily transmit diseases from the mother plants, causing yellow ring spots and artichoke decline.5 The infection of these viruses often severely affects the quality and yield of commercial artichoke plants.5 In Vietnam, five imported artichoke varieties flowered simultaneously at a similar time as the AX variety, approximately seven months after being planted in the field. This study indicated that all five imported artichoke varieties achieved good yield and flower quality, with Artichoke Cardon Blanc Ivoire AB (purple flowers) and Artichoke Green Globe (green flowers) being the two varieties with higher flower yields, as well as unique flower shapes and colors. The remaining varieties had lower yields and were comparable to the domestic AX variety.6
Artichoke is a rich source of polyphenolic compounds, mainly caffeoylquinic acid derivatives and flavonoids.4,7,8 Total polyphenols are more abundant in leaves than in the flower heads, making artichoke leaves an important source of beneficial phenolic compounds for health. The most abundant polyphenols are chlorogenic acid and 1,5-di-O-caffeoylquinic acid (1,5-diCQA).7,8 The predominant flavonoids belong to the flavone class, with scolymoside and cynaroside being the most prevalent. Studies have shown that the n-butanol and water fractions of artichoke leaves possess the highest total polyphenol content and strongest antioxidant activity.9 However, the polyphenol content can vary due to factors such as environmental conditions, genetics, harvesting time, and different plant parts (leaves, flower heads, bracts of flower heads).4,7,8,10,11
In both leaves and flower of artichoke, the main components are mono-caffeoylquinic acids (CQA, 0.48%–4.24%), followed by di-caffeoylquinic acids and flavonoids (0.03%–0.52%).12,13 Some isolated CQA derivatives include 4 mono-caffeoylquinic acid derivatives,7,8,10,14–16 these hydroxycinnamic acids are primarily in the trans configuration, although exposure to ultraviolet radiation may generate cis isomers in plant tissues.10,17 Other phenolic acids identified in artichoke are caffeic acid, p-coumaric acid, ferulic acid, and syringic acid, but they are found in lower concentrations.8,10 Though not a major component, cynarin is highly notable for its various biological effects, such as promoting bile production and protecting the liver.18–22 Some studies suggest that cynarin is not a native constituent of artichoke but rather a compound formed from the isomerization of 1,5-diCQA during high-temperature water extraction.23–27 Consequently, the cynarin content in dried artichoke flower heads or leaves is very low or undetectable.14 However, it can be found in significant amounts in concentrated extracts or products containing artichoke extracts.27
Around 10% of artichoke contains flavonoids, mainly derivatives of luteolin, apigenin, and anthocyanidins (cyanidin, peonidin, and delphinidin). Some flavanone compounds as naringenin and hesperetin have also been identified.23 Flavonoids are present in both leaves and flower heads of artichoke, while anthocyanidins are exclusive to the flower heads. Flavonoid content decreases with the age of the flower, while anthocyanidin content increases in mature flowers.28 The characteristic bitterness of artichoke is mainly attributed to the presence of guaianolide-type sesquiterpene lactones. The two main components found are cynaropicrin and grosheimin. Cynaropicrin is the principal bitter compound in artichoke leaves.29,30 The bitterness is reduced if the structures of these sesquiterpene lactones lose exomethylene or undergo lactone ring opening.29 Cynaropicrin is most abundant in young leaves or leaves at the top of the plant, less in older leaves and bracts, and not detected in outer bracts, roots, mature flowers, and fruit.8,10 Other minor constituents include saponins,31 lignans (derivatives of pinoresinol, including 1-hydroxypinoresinol 1-O-β-D-glucoside, pinoresinol 4-O-β-D-glucoside, pinoresinol acetylhexoside, (+)-pinoresinol),8,10 and essential oils.8,32,33 Artichoke is also rich in vitamins A–C, folic acid, and various amino acids.15,34 The main components identified are β-selinene (40%) and caryophyllene (19%).8,32,33 The edible portion of artichoke flower heads contains a high content of inulin, mainly located in the bracts (about 75% of total carbohydrates).12
Studies conducted in vitro have confirmed the antioxidant properties.9 The antioxidant activity of artichoke depends on the dosage and has been found to be particularly beneficial in treating liver diseases compared to other conditions.35 Artichoke can inhibit lipid peroxidation and neutralize free radicals by acting as a reducing agent, hydrogen donor, and metal chelator. It also regulates ROS-dependent cell signaling at specific key sites.36 This regulatory effect may result from blocking free radicals and ROS-related protein kinases, phosphatases, and various transcription factors.37–41
The 70% ethanol extract of artichoke (at a dosage of 300 mg/kg) and silymarin both significantly reduced liver weight and liver enzymes compared to the control group.42 Artichoke at the same dosage (300 mg/kg) was also shown to decrease ALT, AST, and ALP levels to normal in mouse models of lead toxicity.43,44 The mechanism of action was demonstrated through artichoke’s ability to reduce DNA fragmentation, p53, and caspase 3 levels, thereby aiding in the repair of DNA damage caused by CCl4-induced liver toxicity.45 The hepatoprotective effects against CCl4 toxicity of cynarin, caffeic acid, and cynaroside found in Artichoke have also been reported.46 The leaf extract of artichoke has the ability to inhibit AKR1B1 (an enzyme belonging to the aldo-keto reductase family).47In vivo, the anti-inflammatory effect of artichoke has been demonstrated in experiments inducing mouse paw edema caused by carrageenan. This effect is attributed to luteolin, which has inhibitory activity against COX-2, PGE2, lipopolysaccharide (LPS), xanthine oxidase, lipoxygenase, and TNF-α.48,49 Additionally, artichoke has beneficial effects on the liver, cardiovascular system, and digestive system.50
However, to date, there have been many limitations in studying the chemical composition and biological activity of artichoke plants harvested in Vietnam. In this study, we present the chemical composition and antioxidant activity results using the 1,1-diphenyl-2-picrylhydrazyl radical scavenging and malondialdehyde activity models of artichoke samples harvested in Lam Dong province, Central Vietnam.
Experimental
Plant materials – The leaves of the artichoke (green variety) were harvested in Lam Dong, Vietnam, in October 2020. The identification and scientific naming were carried out by Dr. Vo Van Chi. The plant sample is stored in the Department of Medicinal Plants, Faculty of Pharmacy, University of Medicine and Pharmacy in Ho Chi Minh City.
Instruments and reagents – In the process of extraction and isolation of components, the solvents used, including methanol, chloroform, ethyl acetate, n-butanol, and formic acid, meet the criteria of purity for analysis. For the HPLC analysis were performed on an Alliance 2695 XE HPLC system (Waters, USA) and a Sunfire C18 column (250 × 4.6 mm; 5 μm) (Waters, USA), using acetonitrile (ACN) with 0.1% formic acid 0.1% as elution solvents. The Prominence preparative HPLC system (Shimadzu, Japan) was used to separate individual components with a Sunfire C18 column (150 × 10 mm; 5 μm) (Waters, USA) or a Luna C18 column (250 × 10 mm; 5 μm) (Phenomenex, USA). The solvents used are Acetonitrile (from Scharlau), Formic acid (from Merck), Trifluoroacetic acid (from Prolabo), and twice-distilled water. Pre-coated silica gel F254 plates (0.25 mm thickness, from Merck) are used. Additionally, Silica gel 60 (40–63 μm, Merck), Silica gel C18 (40 × 63 μm), and Sephadex LH-20 are used for isolation purposes. The DPPH test includes DPPH reagent (Sigma, batch number STBD1145V), methanol (Labscan), and DMSO (Merck) as solvents. The MDA test involves 1.15% KCl, 50 mM phosphate buffer at pH 7.4 (KH2PO4, K2HPO4), 10% trichloroacetic acid (TCA), and 0.8% thiobarbituric acid (TBA).
Free Radical Scavenging Activity Assayed by DPPH Test – DPPH (1,1-diphenyl-2-picrylhydrazyl) is a free radical with a maximum absorption wavelength (λmax) of 517 nm and appears purple in color. Antioxidant compounds will neutralize the DPPH free radical by donating hydrogen, reducing the absorption intensity at the maximum wavelength, and causing the solution to change from purple to yellow. The experimental model was conducted by Kulisic et al. (2004) and supplemented by Obeid et al. (2005).51 The applied DPPH concentration was 0.08 mM, the detection wavelength was 517 nm, and the absorbance was measured 30 minutes after the reaction. The experiment was repeated three times, and the average value ± standard deviation (SD) was recorded. Ascorbic acid and silymarin were used as reference substances.
Ex vivo antioxidant activity assayed by the MDA method – Healthy male Swiss albino mice, weighing 25 ± 2 g and aged 5 to 6 weeks, were provided by the Vaccine and Biological Products Medical Institute - Nha Trang, Vietnam. The mice were stably fed with pellet food, vegetables, and water for at least 1 week before the experiment. The mice were sacrificed, and their livers were used for the MDA (malondialdehyde) ex vivo test. MDA (malondialdehyde) is a substance generated during the peroxidation of cellular membrane lipids. When reacted with the thiobarbituric acid (TBA) reagent, it forms a pink-colored trimethine complex with maximum absorption (λmax) at 532 nm. The reaction occurs at pH 2–3, at a temperature of 90–100°C for 10–15 minutes. The antioxidant activity of the test compounds is demonstrated by reducing the color intensity of this complex, indicating a reduction in the amount of MDA present in the sample, leading to reduced absorption at 532 nm. To perform the experiment, first, the mouse livers were separated and homogenized in a 1.15% KCl solution (1 g/10 mL ratio) at temperatures ranging from 0 to 5°C. Fifty microliters of various concentrations of the test sample were mixed with 250 μL of liver homogenate. Then, 1 mL of phosphate buffer (pH = 7.4) was added to the mixture, which was allowed to react for 60 minutes at 37°C. The reaction was stopped by adding 500 μL of 10% trichloroacetic acid (TCA). After centrifugation (3,000 rpm for 10 minutes), 500 μL of the upper solution was taken and reacted with 500 μL of 0.8% thiobarbituric acid at 100°C for 15 minutes. The absorbance at 532 nm was measured after cooling. The measurements were performed using a 96- well plate and an Elisa reader.52 The experiment was conducted twice, and the average value ± standard deviation (SD) was calculated. Silymarin was used as the reference substance. The antioxidant activity (% AO activity) of the test samples was calculated using the following formula:
| (1) |
where OD stands for Optical Density (absorbance). The IC50 of the test sample was determined through regression analysis.
Extraction and isolation – Fresh leaves of Cynara scolymus (90 kg) were washed and steamed at 100°C for 10 minutes. Then, they were extracted using the hot soaking method (80–90°C) for 4 hours with ethanol-water (1:1) (v/v) solution. The extract was concentrated to a thick liquid (5.2 kg) by solvent recovery. Water was added, and the mixture was sequentially shaken with chloroform (4 × 3 liters), ethyl acetate (3 × 3 liters), and n-butanol (3 × 5 liters). The solvent fractions obtained were 60 g of chloroform extract (Cf), 30 g of ethyl acetate extract (EA), and 90 g of n-butanol extract (Bu). Bu extract (10 g) was dissolved in methanol, resulting in precipitation. The precipitate was washed with cold methanol, yielding compound 19 (133.5 mg). The remaining solution was filtered and concentrated. The concentrated solution was dissolved in a minimal amount of distilled water for further separation using MPLC 170 g of C18 silica gel with an elution gradient of acetonitrile (ACN) with 0.1% formic acid. This process resulted in the isolation of 10 fractions (Bu-1 to Bu-10). Fractions Bu-2 (0.35 g) and Bu-3 (0.43 g) were separated using prep-HPLC with a Sunfire Prep C18 column (10 × 150 nm, 5 μm) and eluted with a gradient of ACN (B) with 0.1% FA (A), the composition was changed from 92A:8B (v/v) to 85A:15B (v/v) and 70A:30B (v/v) respectively for 25 min, the flow rate was set at 4 mL/min, and the injection volume 200 μL of each fraction (50 mg/mL); UV detection was performed at 323 nm. This yielded compounds 2 (12.36 mg) and 3 (13.43 mg) from Bu-2, and compounds 4 (49.4 mg) and 5 (21.7 mg) from Bu-3. The amorphous precipitate of fraction Bu-4 was recrystallized in a mixture of methanol and water, resulting in compound 7 (83.5 mg). Fraction Bu-5 (0.083 g) was further purified on a Sunfire Prep C18 column using an elution gradient of ACN with 0.1% FA 0.1% 25 minutes, yielding compound 6 (9.49 mg). The precipitate from Bu-6 was recrystallized in a mixture of CHCl3 and MeOH, resulting in compound 14 (197.8 mg). The precipitate from Bu-7 was filtered and washed with cold methanol, yielding compound 13 (120 mg). Fraction Bu-8 was freeze-dried, resulting in compound 8 (85.2 mg). Fraction Bu-9 (1.5 g) was further separated using a Phenomenex Luna C18 column (10 × 250 mm; 5 μm) and eluted with MeOH (B) and FA 0.1% (A), the composition was changed from 70A:30B (v/v) to 63A:37B (v/v) for 60 min, the flow rate was at 3 mL/min, and the injection volume 100 μL of fraction (50 mg/mL); UV detection was performed at 323 nm. This process yielded compounds 9 (63.3 mg), 10 (74.4 mg), and 15 (2.4 mg), respectively. EA extract (30 g) was isolated using VLC column chromatography (800 g of silica gel, particle size 40–63 μm) with an elution gradient of CHCl3-EtOAc ranging from a ratio of 10:0 to 1:9. This process yielded 16 fractions (EA-1 to EA-16). Fraction EA-2 (1.12 g) was recrystallized in CHCl3-MeOH, resulting in compound 20 (180.7 mg). The precipitate from fraction EA-3 (0.27 g) was filtered and washed with cold CHCl3-MeOH, obtaining compound 12 (16.7 mg). Fraction EA-4 (0.35 g) was recrystallized in MeOH, yielding compound 17 (13.2 mg). Fraction EA-5 (2.4 g) was further separated using MPLC (170 g of C18 silica gel) with an elution gradient of ACN with 0.1% FA. This resulted in corresponding compounds 16 (48.6 mg) and 1 (49.9 mg). Fraction EA-6 (0.32 g) was separated on a Sephadex LH-20 column (100 g) with an elution gradient using 100% MeOH, yielding compound 18 (6.2 mg). Fraction EA-16 (1.2 g) was further separated using MPLC (170 g of C18 silica gel) with an elution gradient of ACN with 0.1% FA. This process yielded corresponding compounds 8 (35.2 mg) and 11 (43.1 mg).
Statistical analysis – The data are presented as mean values ± standard deviation (SD) and were statistically analyzed using the One-Way ANOVA method and the Tukey test with Minitab 19 software. Differences between the mean values are considered statistically significant when the p-value is less than 0.05 at a 95% confidence level.
Results and Discussion
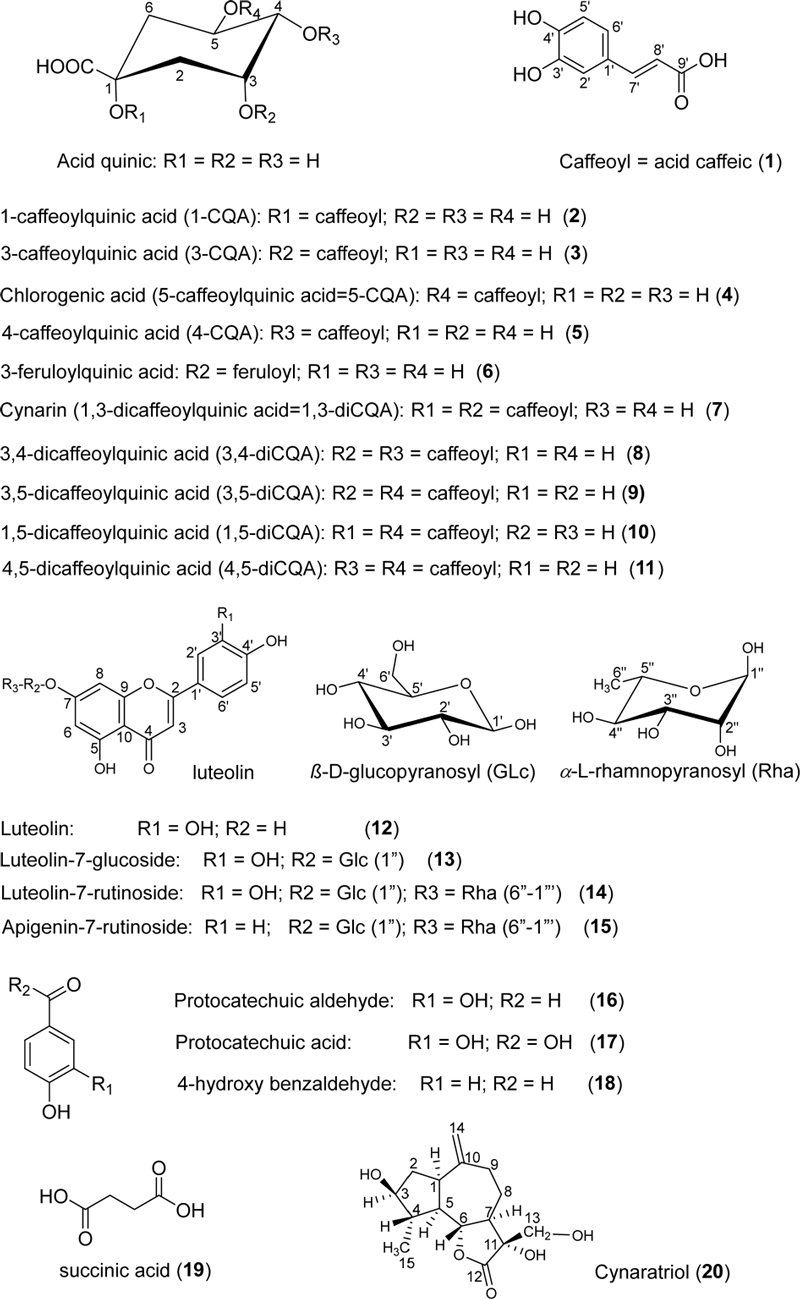
Among the 20 compounds extracted from artichoke leaves, eleven compounds are derivatives of caffeic acid (1–11). These compounds are white, amorphous powders that dissolve in methanol and yield a positive reaction with the FeCl3 reagent. Their UV spectra (λmax in MeOH) exhibit three similar absorption peaks at 217.9 nm, 240.2 nm, and 324.4 nm. However, compound 7 displays a slightly different shoulder shape in the UV spectrum compared to the others, with peaks at 216.8 nm, 243.8 nm, and 323.1 nm. Based on the characteristic fragmentation patterns observed in the ESI-MS spectra (Supporting information S1, 2), these compounds were identified as derivatives of caffeic acid.53 Among them, compound 1 exhibited [M−H]– at m/z 179.1, corresponding to [caffeoyl-H]–, indicating a molecular weight of 180 and a predicted molecular formula of C9H8O4. Compounds 2–6 are predicted to be mono-CQA as they all display [M−H]– ions at m/z = 353.1 and characteristic fragment ions at m/z = 191.1 [quinic acid−H]– or [M−caffeoyl− H]–, and m/z 179.1 [caffeoyl−H]–. These compounds were suggested to contain one quinic acid and one caffeic acid moiety. Interestingly, compound 6 with m/z = 367.2 exhibits similar fragment ions as 1–5, resulting in a calculated molecular formula of C17H20O9 (367 Da) (Supporting information S1-5). However, compounds 7–11 display [M−H]– ions at m/z = 515.1 and characteristic ions including m/z = 353.1 [M− caffeoyl−H]–, these compounds are predicted to be di-CQA with a calculated molecular formula of C25H24O12 which content a quinic acid and two caffeic acid moieties.
In the 1H NMR spectrum, most of the caffeic acid moiety in 1–11 were identified based on the appearance of two doublet peak pairs of two protons at δH 7.37–7.55 ppm (J = 16.0 Hz) and 6.11–6.28 (J = 16.0 Hz), indicative of two olefinic protons H-7′(H-7′′) and H-8ʹ(H-8′′) with a trans relationship to each other in one or two caffeic acid groups. Additionally, the three proton signals of the aromatic ring interacted with each other in an ABX system [H-2ʹ(H-2′′) d (2.0 Hz); H-5′(H-5′′) d (8.0 Hz); H-6′(H-6′′) dd (2.0 and 8.0 Hz)], indicating that the aromatic ring has three substituents at positions 1, 3, and 4.54
In the 13C NMR spectrum, compound 1 exhibits 9 carbon signals, including 5 carbon signals of CH groups and 4 quaternary carbons [a carbonyl group at δC 165.3–168.7 ppm (C-9ʹ); two oxymethine carbons in the aromatic ring at δC 145.1–149.5 (C-3ʹ and C-4ʹ); and one remaining signal at δC 125.3–127.8 (C-1′)] (Table 3). Compounds 2–6 display 16 carbon signals, including 9 signals similar to 1, and 7 additional signals [a signal at δC 171.1–177.1 (C-7) of the carbonyl group and 6 signals in the high field region at δC 73.1–81.2]. Compound 6 exhibits an additional strong signal at δC 51.7, indicating a methoxy group. Meanwhile, 7–11 contain 25 carbon signals, including 16 signals similar to 2–6, and 9 additional signals similar to 1. The quinic acid moiety in these compounds was identified based on the 7 signals in the 13C-NMR spectrum [three oxymethine carbons at δC 63.9–77.0 (C-3, C-4, and C-5), two methylene carbons at δC 33.0–40.8 (C-2 and C-6), one oxidized quaternary carbon at δC 72.9–81.2 (C-1), and one carbonyl group at δC 173.1–178.2 (C-7)] (Tables 3 and 4, and Supporting information S6-15).
Normally, the stereochemistry of quinic acid is determined based on the coupling pattern of J3,4 and J4,5.53 The coupling constants of 2–11 were J3,4 (3.0–3.5 Hz) and J4,5 (8.0–11.5 Hz), indicating that their stereochemical configurations are consistent with the signals of H-3 (equatorial), H-4 (axial), and H-5 (axial).55,56 Analysis of the proton signals of quinic acid in the high field region at δH 3.31–5.63 (H-3, H-4, and H-5) by observing the downfield signals of these protons compared to the spectrum of quinic acid demonstrates that these positions are linked to the acyl groups (Table 1 and Table 2).53,55,56 The chemical structures of these compounds were also confirmed by 2D NMR including COSY and HMBC spectrum (S6-15), which provide interactions between (H-2′ and C-7′); (H-7′ and C-9′) to confirm the linkage of the aromatic ring with the C3 of olefinic side chain. Therefore, these compounds were identified as 1-caffeoylquinic acid (2), 3-caffeoylquinic acid (3), 5-caffeoylquinic acid (chlorogenic acid) (4), 4-caffeoylquinic acid (5), and 3-feruloylquinic acid (6), 1,3-di-caffeoylquinic acid (cynarin) (7), 3,4-di-caffeoylquinic acid (8), 3,5-di-caffeoylquinic acid (9), 1,5-di-caffeoylquinic acid (10), and 4,5-di-caffeoylquinic acid (11).53,55,56
Among the isolated metabolites, 12–15 were identified as flavonoid. These compounds are yellow amorphous powders, soluble in methanol, ethanol, and ethyl acetate, and show positive reactions with FeCl3 and AlCl3 reagents. The UV spectra (λmax MeOH) of these compounds exhibit three absorption peaks at 211.7, 254.3, and 347.2 nm. Their ESI-MS (ES)– spectra (m/z) (S3) are analyzed as 12 (m/z 285.1 [M–H]–), 13 (m/z 447.6 [M–H]–, 285.5 ([M–glc–H]–), 14 (m/z 593.3 [M–H]–; 447.4 [M–rh –H]–), and 15 (m/z 577.0 [M–H]–; 269.3 [M–glc–rha–H]–)
In the 1H-NMR spectrum, signals at δH 6.18–7.80 ppm shows that compounds 12–14 contain 5 aromatic proton signals including two protons at δH 6.1–6.79 (1H, d, 2.0) belonging to ring A, and 3 protons belonging to ring B of the flavone framework at δH 7.39–7.42 (1H, d, 2.0), δH 6.87–6.93 (1H, d, 8.5), and δH 7.39–7.80 (1H, dd, 8.5, 2.0)57,58 (Table 5). These protons interact with each other in an ABX system, indicating that ring B is substituted at positions 1ʹ , 3ʹ, and 4ʹ. Compound 15 has similar ring A signals to 12–14, but ring B of 15 has two pairs of signals at δH 6.87 (2H, d, 8.0) and δH 7.80 (2H, d, 8.0), indicating 2 symmetrically equivalent CH protons in the aromatic ring, and thus, 15 has one OH group in ring B (see Supporting information S16-19).
In the 13C-NMR spectrum, signals for 15 carbon atoms in the range of δC 98.9–184.0 ppm, comprising 6 tertiary carbons, 8 quaternary carbons, and 1 carbonyl group were observed (Table 5). All tertiary and quaternary carbons manifest in the low-field region typical of sp2 carbon signals found in double bonds within aromatic rings. A distinctive coupling constant signal in the low-field region at δC 181.7–184.0 (C-4) is indicative of a flavonoid framework.57 Specifically, 15 exhibits two carbon signals at δC 115.7 (C-2ʹ and 6ʹ) and δC 128.3 (C-3ʹ and 5ʹ), suggesting a symmetrically structured ring B with only one hydroxy group connecting to C-4′ (Table 5 and Supporting information S19). Notably, compounds 13–15 display additional carbon signals at δC 60.6–77.8, indicating the presence of sugar moieties. Compound 13 is identified as a mono-glycoside, while compounds 14–15 are di-glycosides due to the presence of more than four coupling signals in this region compared to compound 12. These findings align with the signals observed in the MS spectra of these compounds (see Supporting information S3). Furthermore, the sugar moieties of 13–15 are determined based on the coupling signals of protons at δH 3.27-5.08 ppm in the 1H-NMR spectrum (Table 5 and Supporting information S16-19). These compounds exhibit doublet signals at δH 4.96–5.08 (1H, d, 7.5), indicative of an O-β-D-glycosidic linkage, with the coupling constant of 7.5 Hz. Additionally, the 1H-NMR data, combined with five coupling signals at δC 60.1–77.9 and one signal at δC 99.9–101.7, suggest the presence of β-D-glucopyranose. The second sugar moiety of 14 and 15 is identified as α-L-rhamnopyranose based on the proton signal at δH 4.62 (1H, d, 1.5) or a broad singlet (1H, s, brd), along with a doublet signal of the methyl group at δH 1.08–1.21 (1H, d, 6.4) (Supporting information S18, 19). The linkage position of the sugar moiety at C-7 of 13–15 is confirmed by the interaction of the anomeric proton (H-1′) of the sugar with this carbon in the HMBC spectrum. Furthermore, the interaction between the signal at δH 4.74–4.62 (1H, d, 1.5, H-1′′′) and δC 66.0–67.5 (C-6′′) allows for the determination of the linkage between the two sugars as (1→6). Comparing the 1H-NMR and 13C-NMR spectra with the published data, these compounds were identified as and luteolin (12), luteolin-7-O-[β-D-glucopyranosyl-(6→1)-α-L-rhamnopyranosyl] or luteolin-7-O-rutinoside (scolymoside) (14),58 luteolin-7-O-β-D-glucopyranoside (cynaroside) (13),59 apigenin-7-O-[β-D-glucopyranosyl-(6→1)-α-L-rhamnopyranosyl] or apigenin-7-O-rutinoside (isorhoifolin) (15).60,61
Compound 16 presents as a colorless amorphous powder, soluble in methanol, and demonstrates a positive reaction with the FeCl3 reagent. The UV spectrum exhibits three absorption peaks at 230.8, 279.2, and 311.3 nm in methanol solution. In the ESI-MS spectrum (ES–), the molecular ion peak [M−H]– appears at m/z 137, suggesting a molecular formula of C7H6O3. The 1H NMR spectrum reveals three proton signals at δH 7.22–7.32 ppm (1H, d, 2.0), 6.73–6.89 (1H, d, 8.0), and 7.25–7.25 (1H, dd, 8.0, 2.0), indicating an ABX spin system, implying substitution at positions 1, 3, and 4 of the aromatic ring. The 13C NMR spectrum displays seven carbon signals, including one carbonyl at δC 168.4 (C-7) (Supporting information S20). Based on the NMR spectrum and comparison with published data, the compound is identified as aldehyde protocatechuic.62
Compound 17 was isolated as a colorless amorphous powder, soluble in methanol, and exhibits a positive reaction with the FeCl3 reagent. The UV spectrum reveals three absorption peaks at 220.2, 259.1, and 293.5 nm in methanol solution. In the ESI-MS spectrum (ES–), the molecular ion peak [M−H]– appears at m/z 153.0, suggesting a molecular formula of C7H6O4. The 1H NMR spectrum displays three proton signals at δH 7.22–7.32 ppm (1H, d, 2.0), 6.73–6.89 (1H, d, 8.0), and 7.2-7.25 (1H, dd, 8.0, 2.0), indicative of an ABX spin system, implying substitution at positions 1, 3, and 4 of the aromatic ring. The 13C NMR spectrum exhibits seven carbon signals, including one carboxylic acid group at δC 173.5 (C-7) (see Supporting information S21). Based on the NMR spectrum and comparison with published data, the compound is identified as acid protocatechuic.62
About the compound 18, in the ESI-MS spectrum (ES–), the molecular ion peak [M−H]– appears at m/z 121.1, indicating a molecular formula of C7H6O2. The 1H-NMR spectrum shows two proton signals at δH 6.80 ppm (2H, d, 8.0), and 7.77 (2H, d, 8.0), suggesting a symmetric structure. The 13C-NMR spectrum displays seven carbon signals, including two strong signals at δC 131.4 (C-2 and C-6) and 115.0 (C-3 and C-5) (Table 6), indicating a symmetric aromatic ring (see Supporting information S22). Based on the NMR data and comparison with published data, 18 is identified as 4-hydroxybenzaldehyde.62
Compound 19 is a colorless crystalline solid with a sour taste and good solubility in water. In the APCI-MS (APCI–) spectrum, 19 shows an ion [M−H]– with m/z at 117.0. Therefore, the molecular formula is predicted to be C4H6O4. In the 13C and 1H NMR spectra (see Supporting information S23), 19 exhibits only 2 strong carbon signals, indicating a symmetrical structure. These signals correspond to one carbon of the carboxylic acid group at δC 173.5 ppm and one carbon of the methylene group at 28.8 ppm. In the 1H NMR spectrum, a signal at δH 12.1 (1H, s) is assigned to the proton of the carboxylic acid group, and a strong signal at δH 2.42 (2H, s) is assigned to the protons of the methylene group. Based on the data from the MS and NMR spectra, 19 is identified as succinic acid.63
Compound 20 is a colorless needle that is soluble in chloroform but poorly soluble in methanol. It lacks any UV absorption at 254 and 365 nm, but it produces a purple color with the vanillin-sulfuric acid (VS) reagent. The APCI-MS spectrum displays a molecular ion peak at m/z 283.3, corresponding to molecular formula of C15H22O5. In the 1H-NMR spectrum, 20 exhibits signal for two proton methylenes at δH 3.42–4.82 ppm (2H, m) attributed to the protons of the oxymethylene and olefinic methylene groups. Additionally, three signals for other proton methylenes are observed at δH 1.49–1.94 (3H, m). Six signals for proton methines are present, with two signals at δH 3.42–3.98 (1H, m) assigned to the protons of the oxymethine group, and the remaining four signals at higher field strength at δH 1.49–2.69 (1H, m) assigned to non-oxidized methine protons (Supporting information S24). The 13C-NMR spectrum of 20 displays 15 carbon signals, including three quaternary carbon groups, one of which is a carbonyl group at δC 178.6 ppm, six tertiary carbons, five methylene carbon groups, and one methyl group. The signals for tertiary carbons in the high field range at δC 76.86–83.75 are characteristic of the oxymethine carbons. Additionally, one signal for a secondary carbon is observed at low field at δC 111.43, assigned to the olefinic methylene carbon, and one signal at δC 63.12 is assigned to the oxymethylene carbon. Based on the 13C-NMR and the overall chemical composition, 20 is predicted to be a sesquiterpene lactone belonging to the guaianolide skeleton (Supporting information S24). The interactions observed in the COSY spectrum between the proton of the methine and methylene groups aid in determining their positions in the cyclopentane and cycloheptane rings of the guaianolide skeleton. Additionally, the HMBC spectrum reveals interactions between the hydroxy group protons and corresponding carbons, as well as interactions between olefinic and ketone groups with their respective protons, further supporting the confident determination of their positions in the structure. Therefore, the structure of 20 is confirmed to be cynaratriol.64
The investigation into antioxidant activity focused on two main mechanisms: the ability to neutralize free radicals through hydroxyl group donation and the capability to reduce metal ions. Out of the 20 isolated compounds, nine polyphenolic compounds sourced from artichoke leaves were assessed for their antioxidant activity using DPPH radical scavenging activity, owing to limitations in sample quantities.
The antioxidant activity assessment of the nine purified polyphenolic compounds isolated from artichoke leaves revealed that all tested compounds as 1-CQA (2), 3-CQA (3), chlorogenic acid (4), 4-CQA (5), cynarin (7), 1,5-diCQA (10), 4,5-diCQA (11), cynaroside (13), and scolymoside (14) displayed robust DPPH radical scavenging activity, with IC50 values ranging from 2.6 to 21.7 μg/mL (Table 7). Notably, the flavonoid compounds (cynaroside and scolymoside) and di-CQA compounds (cynarin; 1,5-diCQA; 4,5-diCQA) exhibited particularly significant activity, with IC50 values ranging from 5.7 to 7.7 μM, surpassing the potency of vitamin C (33.5 μM). The ranking of antioxidant activity among these compounds is as follows: cynaroside > 4,5-diCQA > 1,5-diCQA = cynarin-1,3-diCQA = scolymoside > 4-CQA > chlorogenic acid > vitamin C > 3-CQA > 1-CQA > silymarin.
The antioxidant activity of these polyphenolic compounds may involve various mechanisms, including Hydrogen Atom Transfer (HAT), Single Electron Transfer (SET), and the ability to form chelate complexes with Fe2+. These effects are closely linked to their structural characteristics, particularly the number and arrangement of hydroxyl groups on the aromatic rings.63 Furthermore, the antioxidant activity is largely influenced by the number of hydroxyl groups on the aromatic rings. Generally, a higher number of hydroxyl groups corresponds to stronger antioxidant activity. Additionally, the presence of a second hydroxyl group at ortho or para positions further enhances antioxidant activity.63
In this experiment, compounds like cynarin and its isomers di-CQA, alongside flavonoid derivatives of luteolin, are noted for possessing two hydroxyl groups on their aromatic rings. This feature contributes to their stronger antioxidant activity compared to chlorogenic acid or other mono-CQAs (1-CQA, 3-CQA, and 4-CQA) with two hydroxyl groups adjacent on the same aromatic ring. Moreover, cynaroside, a flavonoid with fewer sugar molecules in its structure compared to scolymoside, demonstrates stronger antioxidant activity than scolymoside (which contains two sugar molecules). This distinction is evident from the IC50 values, where cynaroside exhibits an IC50 of 5.51 μM compared to 7.79 μM for scolymoside. Regarding the antioxidant activity of di-CQAs (such as cynarin or 1,3-diCQA; 1,5-diCQA; 3,4-diCQA; 3,5-diCQA; 4,5-diCQA), a study by Li et al. revealed that di-CQAs with two adjacent caffeoyl groups (4,5-CQA and 3,4-CQA) consistently exhibit higher antioxidant activity than non-adjacent di-CQAs (1,3-CQA; 1,5-CQA; and 3,5-CQA).64 However, 1,3-CQA and 1,5-CQA demonstrate better cell protection than adjacent di-CQAs. These differences could be attributed to the position of the two caffeoyl groups and their structural effects on the quinic acid framework.
Several proposed mechanisms account for the variation in antioxidant activity of di-CQAs. Firstly, 4,5- and 3,4-diCQA contain two adjacent caffeoyl groups, which are close to each other, resulting in a denser molecular structure and increased molecular energy, leading to enhanced redox reactions.64 Therefore, in the DPPH-based antioxidant activity assay that relies on redox reactions, 4,5- and 3,4-diCQA exhibit stronger activity than non-adjacent di- CQAs (1,3-CQA, 1,5-CQA, and 3,5-CQA). Conversely, cynarin (1,3-diCQA) and 1,5-diCQA have two distant caffeoyl groups on the quinic acid framework, leading to reduced molecular energy and diminished redox capacity. Secondly, the antioxidant activity of polyphenols is also demonstrated through their ability to form chelate complexes with Fe2+ using the two hydroxyl groups of the caffeoyl framework. Two adjacent caffeoyl groups in 3,4-CQA and 4,5-CQA extend from the same side of the quinic acid framework, enabling them to better surround Fe2+ in chelate reactions. In contrast, the two caffeoyl moieties in non-adjacent di-CQAs (1,3-CQA and 1,5-CQA) extend from different directions, making it difficult for them to effectively surround Fe2+ for participation in the reaction. Therefore, in the Ferric Reducing Antioxidant Power (FRAP) assay, these compounds usually have weaker antioxidant activity than adjacent di-CQAs. This mechanism is supported by the complex formation assay with Fe2+, where 3,4- and 3,5-diCQA showed deeper blue color and higher UV absorption than 1,5- and 1,3-diCQA. Regarding the antioxidant activity using the MDA model, 1,3- di-caffeoylquinic acid (or cynarin) (7) showed the strongest MDA activity with an IC50 value of 24.7 μM, followed by 1,5-diCQA (66.8 μM), and 4,5-diCQA (127.3 μM). The other compounds showed weak inhibitory activity with IC50 values over 150 μM.
This study aimed to identify the chemical compounds present in artichoke plants harvested in Lam Dong province, Vietnam, and assess their antioxidant capabilities using two research models: DPPH and MDA. The DPPH model evaluates the free radical scavenging ability, while the MDA model assesses the capacity to reduce malondialdehyde levels, a substance produced during lipid oxidation. Though facing limitations, the study successfully isolated 20 phenolic compounds (1–20) and evaluated the antioxidant activity of 9 compounds using the DPPH and MDA models. By publishing these research findings, the aim is to pave the way for optimal utilization of artichoke resources in Vietnam and contribute to a better understanding of the biological capabilities and potential applications of this plant in healthcare and human well-being.
In conclusion, our study provides comprehensive information on the isolation and identification of 20 phenolic compounds from Vietnamese artichoke, including caffeic acid (1), 1-caffeoylquinic acid (2), 3-caffeoylquinic acid (3), 5-caffeoylquinic acid (chlorogenic acid) (4), 4-caffeoylquinic acid (5), 3-feruloylquinic acid (6), 1,3-di-caffeoylquinic acid (cynarin) (7), 3,4-di-caffeoylquinic acid (8), 3,5-di-caffeoylquinic acid (9), 1,5-di-caffeoylquinic acid (10), and 4,5-di-caffeoylquinic acid (11), as well as luteolin (12), luteolin-7-O-β-D-glucopyranoside (cynaroside) (13), luteolin-7-O-[β-D-glucopyranosyl-(6→1)-α-L-rhamnopyranosyl] or luteolin-7-O-rutinoside (scolymoside) (14), apigenin-7-O-[β-D-glucopyranosyl-(6→1)-α-L-rhamnopyranosyl] or apigenin-7-O-rutinoside (isorhoifolin) (15), protocatechuic acid (16), protocatechuic acid (17), 4-hydroxybenzaldehyde (18), succinic acid (19), and cynaratriol (20). Furthermore, our study also demonstrates the antioxidant activity of the 9 purified polyphenolic compounds using both the DPPH and MDA tests. This result contributes to providing a database of the antioxidant activity of artichoke extracts and sheds light on the structure-activity relationship as well as the correlation between the total polyphenolic content and antioxidant activity in artichoke extracts.
Acknowledgments
This study was supported by University of Medicine and Pharmacy at Ho Chi Minh City (NTMN-2020).
Conflicts of Interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
References
-
Gostin, A.-I.; Waisundara, V. Y. Trends Food Sci. Technol. 2019, 86, 381–391.
[https://doi.org/10.1016/j.tifs.2019.02.015]

- Food and Agriculture Organization of the United Nations, Artichoke. https://www.fao.org/faostat/en/, , 2022 (accessed 1 July 2022).
-
Khai, H. D.; Mai, N. T. N.; Anh, H. L. L.; Nguyet, N. N. M.; Long, H. V.; Luan, V. Q.; Hien, V. T.; Tung, H. T.; Van Lich, T.; Nhung, T. T.; Ha, C. D.; Van Thuc, L; Nhut, D. H. J. Biotechnol. 2021, 19, 129–145.
[https://doi.org/10.15625/1811-4989/15204]

-
De Falco, B.; Incerti, G.; Amato, M.; Lanzotti, V. Phytochem. Rev. 2015, 14, 993–1018.
[https://doi.org/10.1007/s11101-015-9428-y]

-
Minutillo, S. A.; Mascia, T.; Gallitelli, D. Eur. J. Plant Pathol. 2012, 134, 459–465.
[https://doi.org/10.1007/s10658-012-0032-3]

- Khai, H. D.; Cuong, D. M. ; Tung, H. T.; Vinh, N. Q.; Dung, D. M.; Nam, N. B.; Thuc, L. V.; Luan, V. Q.; Mai, N. T. N.; Nhut, D. T. Vietnam J. Sci. Technol. 2021, 64, 37–42.
-
Farag, M. A.; El-Ahmady, S. H.; Elian, F. S.; Wessjohann, L. A. Phytochemistry 2013, 95, 177–187.
[https://doi.org/10.1016/j.phytochem.2013.07.003]

-
Lim, T. K. Edible Medicinal and Non-Medicinal Plants; Springer; Dordrecht, 2013; pp 291–328.
[https://doi.org/10.1007/978-94-007-7395-0_20]

-
Yang, M.; Ma, Y.; Wang, Z.; Khan, A.; Zhou, W.; Zhao, T.; Cao, J.; Cheng, G.; Cai, S. Ind. Crops Prod. 2020, 143, 111433.
[https://doi.org/10.1016/j.indcrop.2019.05.082]

-
Portis, E.; Acquadro, A.; Lanteri, S. The Globe Artichoke Genome; Springer; Cham, 2019; pp 99–113.
[https://doi.org/10.1007/978-3-030-20012-1]

-
Lombardo, S.; Pandino, G.; Mauromicale, G.; Knodler, M.; Carle, R.; Schieber, A. Food Chem. 2010, 119, 1175–1181.
[https://doi.org/10.1016/j.foodchem.2009.08.033]

-
Lattanzio, V.; Kroon, P. A.; Linsalata, V.; Cardinali, A. J. Funct. Foods 2009, 1, 131–144.
[https://doi.org/10.1016/j.jff.2009.01.002]

-
Hausler, M.; Ganzera, M.; Abel, G.; Popp, M.; Stuppner, H. Chromatographia 2002, 56, 407–411.
[https://doi.org/10.1007/BF02492002]

-
Mena-Garcia, A.; Ruiz-Matute, A. I.; Soria, A. C.; Sanz, M. L. J. Chromatogr. A 2021, 1647, 462102.
[https://doi.org/10.1016/j.chroma.2021.462102]

-
Orlovskaya, T. V.; Luneva, I. L.; Chelombit’Ko, V. A. Chem. Nat. Compd. 2007, 43, 239–240.
[https://doi.org/10.1007/s10600-007-0093-2]

-
Panizzi, L.; Scarpati, M. L. Nature 1954, 174, 1062–1062.
[https://doi.org/10.1038/1741062a0]

-
Towers, G. H. N.; Abeysekera, B. Phytochemistry 1984, 23, 951–952.
[https://doi.org/10.1016/S0031-9422(00)82589-3]

-
Fritsche, J.; Beindorff, C. M.; Dachtler, M.; Zhang, H.; Lammers, J. G. Eur. Food Res. Technol. 2002, 215, 149–157.
[https://doi.org/10.1007/s00217-002-0507-0]

- Jun, N.-J.; Jang, K.-C.; Kim, S.-C.; Moon, D.-Y.; Seong, K.-C.; Kang, K.-H.; Tandang, L.; Kim, P.-H.; Cho, S.-M. K.; Park, K.-H. J. Appl. Biol. Chem. 2007, 50, 244–248.
-
Tong, J.; Mo, Q.-G.; Ma, B.-X.; Ge, L.-L.; Zhou, G.; Wang, Y.-W. Food Funct. 2017, 8, 209–219.
[https://doi.org/10.1039/C6FO01531J]

-
Topal, M.; Gocer, H.; Topal, F.; Kalin, P.; Kose, L. P.; Gulcin, İ .; Cakmak, K. C.; Kucuk, M.; Durmaz, L.; Goren, A. C.; Alwasel, S. H. J. Enzyme Inhib. Med. Chem. 2016, 31, 266–275.
[https://doi.org/10.3109/14756366.2015.1018244]

-
Xia, N.; Pautz, A.; Wollscheid, U.; Reifenberg, G.; Forstermann, U.; Li, H. Molecules 2014, 19, 3654–3668.
[https://doi.org/10.3390/molecules19033654]

-
Schutz, K.; Kammerer, D.; Carle, R.; Schieber, A. J. Agric. Food Chem. 2004, 52, 4090–4096.
[https://doi.org/10.1021/jf049625x]

- Wichtl, M. Herbal Drugs and Phytopharmaceuticals: A Handbook for Practice on a Scientific Basis; CRC Press; Marburg, 2004; p 173.
-
Ferracane, R.; Pellegrini, N.; Visconti, A.; Graziani, G.; Chiavaro, E.; Miglio, C.; Fogliano, V. J. Agric. Food Chem. 2008, 56, 8601–8608.
[https://doi.org/10.1021/jf800408w]

- Lutz, M .; H enriquez, C.; E sc obar, M . J. Food Compost. Anal. 2011, 24, 49–54.
-
Schutz, K.; Muks, E.; Carle, R.; Schieber, A. J. Agric. Food Chem. 2006, 54, 8812–8817.
[https://doi.org/10.1021/jf062009b]

-
Lattanzio, V.; van Sumere, C. F. Food Chem. 1987, 24, 37–50.
[https://doi.org/10.1016/0308-8146(87)90082-3]

-
Cravotto, G.; Nano, G. M.; Binello, A.; Spagliardi, P.; Seu, G. J. Sci. Food Agric. 2005, 85, 1757–1764.
[https://doi.org/10.1002/jsfa.2180]

-
Eljounaidi, K.; Comino, C.; Moglia, A.; Cankar, K.; Genre, A.; Hehn, A.; Bourgaud, F.; Beekwilder, J.; Lanteri, S. Plant Sci. 2015, 239, 128–136.
[https://doi.org/10.1016/j.plantsci.2015.07.020]

-
Shimizu, S.; Ishihara, N.; Umehara, K.; Miyase, T.; Ueno, A. Chem. Pharm. Bull. 1988, 36, 2466–2474.
[https://doi.org/10.1248/cpb.36.2466]

-
Buttery, R. G.; Guadagni, D. G.; Ling, L. C. J. Agric. Food Chem. 1978, 26, 791–793.
[https://doi.org/10.1021/jf60218a041]

-
MacLeod, A. J.; Pieris, N. M.; de Troconis, N. G. Phytochemistry 1982, 21, 1647–1651.
[https://doi.org/10.1016/S0031-9422(82)85033-4]

- El Sohaimy, S. A. African J. Food Sci. Technol. 2013, 4, 182–187.
-
Salekzamani, S.; Ebrahimi‐Mameghani, M.; Rezazadeh, K. Phytother. Res. 2019, 33, 55–71.
[https://doi.org/10.1002/ptr.6213]

-
Piston, M.; M ac hado, I .; Branco, C . S.; C esio, V.; Heinzen, H .; Ribeiro, D.; Fernandes, E.; Chiste, R. C.; Freitas, M. Food Res. Int. 2014, 64, 150–156.
[https://doi.org/10.1016/j.foodres.2014.05.078]

-
Leonarduzzi, G.; Sottero, B.; Poli, G. Pharmacol. Ther. 2010, 128, 336–374.
[https://doi.org/10.1016/j.pharmthera.2010.08.003]

-
Brown, J. E.; Rice-Evans, C. A. Free Radic. Res. 1998, 29, 247–255.
[https://doi.org/10.1080/10715769800300281]

-
Jimenez-Escrig, A.; Dragsted, L. O.; Daneshvar, B.; Pulido, R.; Saura-Calixto, F. J. Agric. Food Chem. 2003, 51, 5540–5545.
[https://doi.org/10.1021/jf030047e]

-
Rezazadeh, K.; Rahmati‐Yamchi, M.; Mohammadnejad, L.; Ebrahimi‐Mameghani, M.; Delazar, A. Phytother. Res. 2018, 32, 84–93.
[https://doi.org/10.1002/ptr.5951]

-
Skarpanska-Stejnborn, A.; Pilaczynska-Szczesniak, L.; Basta, P.; Deskur-Smielecka, E.; Horoszkiewicz-Hassan, M. Int. J. Sport Nutr. Exerc. Metab. 2008, 18, 313–327.
[https://doi.org/10.1123/ijsnem.18.3.313]

-
Elgarawany, G. E.; Abdou, A. G.; Taie, D. M.; Motawea, S. M. J. Immunoassay Immunochem. 2020, 41, 84–96.
[https://doi.org/10.1080/15321819.2019.1692029]

-
Heidarian, E.; Rafieian-Kopaei, M. Pharm. Biol. 2013, 51, 1104–1109.
[https://doi.org/10.3109/13880209.2013.777931]

- Heidarian, E.; Soofiniya, Y. J. Med. Plant Res. 2011, 5, 2717–2723.
-
Colak, E.; Ustuner, M. C.; Tekin, N.; Colak, E.; Burukoglu, D.; Degirmenci, I.; Gunes, H. V. SpringerPlus 2016, 5, 1–9.
[https://doi.org/10.1186/s40064-016-1894-1]

-
Xiang, T.; Xiong, Q.-B.; Ketut, A. I.; Tezuka, Y.; Nagaoka, T.; Wu, L.-J.; Kadota, S. Planta Med. 2001, 67, 322–325.
[https://doi.org/10.1055/s-2001-14337]

-
Milačkova, I.; Kapustova, K.; Mučaji, P.; Hošek, J. Phytother. Res. 2017, 31, 488−496.
[https://doi.org/10.1002/ptr.5774]

- Gabriel K, H.; Yong, Q.; Stephen S, L.; Deborah C, S.; Xianglin, S. J. Nutr. 2006, 136, 1517−1521.
-
Wauquier, F.; Boutin-Wittrant, L.; Viret, A.; Guilhaudis, L.; Oulyadi, H.; Bourafai-Aziez, A.; Charpentier, G.; Rousselot, G.; Cassin, E.; Descamps, S.; Roux, V.; Macian, N.; Pickering, G.; Wittrant, Y. Nutrients 2021, 13, 2653.
[https://doi.org/10.3390/nu13082653]

-
Emendorfer, F.; Emendorfer, F.; Bellato, F.; Noldin, V. F.; Cechinel-Filho, V.; Yunes, R. A.; Delle Monache, F.; Cardozo, A. M. Biol. Pharm. Bull. 2005, 28, 902−904.
[https://doi.org/10.1248/bpb.28.902]

-
Kulisic, T.; Radonic, A.; Katalinic, V.; Milos, M. Food Chem. 2004, 85, 633−640.
[https://doi.org/10.1016/j.foodchem.2003.07.024]

-
Ohkawa, H.; Ohishi, N.; Yagi, K. Anal. Biochem. 1979, 95, 351−358.
[https://doi.org/10.1016/0003-2697(79)90738-3]

-
Clifford, M. N. Phytochemistry 1986, 25, 1767−1769.
[https://doi.org/10.1016/S0031-9422(00)81260-1]

-
Lim, E.-K.; Higgins, G. S.; Li, Y.; Bowles, D. J. Biochem. J. 2003, 373, 987−992.
[https://doi.org/10.1042/bj20021453]

-
Horman, I.; Badoud, R.; Ammann, W. J. Agric. Food Chem. 1984, 32, 538−540.
[https://doi.org/10.1021/jf00123a030]

-
Slacanin, I.; Marston, A.; Hostettmann, K.; Guedon, D.; Abbe, P. Phytochem. Anal. 1991, 2, 137−142.
[https://doi.org/10.1002/pca.2800020310]

-
Coxon, B. Anal. Chem. 1983, 55, 2361−2366.
[https://doi.org/10.1021/ac00264a036]

-
Kim, N. M.; Kim, J.; Chung, H. Y.; Choi, J. S. Arch. Pharm. Res. 2000, 23, 237−239.
[https://doi.org/10.1007/BF02976451]

-
Zhu, X. F.; Zhang, H. X. Chem. Nat. Compd. 2004, 40, 600−601.
[https://doi.org/10.1007/s10600-005-0049-3]

-
Kasaj, D.; Krenn, L.; Reznicek, G.; Prinz, S.; Hufner, A.; Kopp, B. Sci. Pharm. 2001, 69, 75−84.
[https://doi.org/10.3797/scipharm.aut-01-09]

-
Owen, R. W.; Haubner, R.; Mier, W.; Giacosa, A.; Hull, W. E.; Spiegelhalder, B.; Bartsch, H. Food Chem. Toxicol. 2003, 41, 703−717.
[https://doi.org/10.1016/S0278-6915(03)00011-5]

-
Lee, I.-C.; Bae, J.-S.; Kim, T.; Kwon, O. J.; Kim, T. H. J. Korean Soc. Appl. Biol. Chem. 2011, 54, 811−816.
[https://doi.org/10.1007/BF03253166]

-
Wang, M.; Simon, J. E.; Aviles, I. F.; He, K.; Zheng, Q.-Y.; Tadmor, Y. J. Agric. Food. Chem. 2003, 51, 601−608.
[https://doi.org/10.1021/jf020792b]

-
Li, X.; Li, K.; Xie, H.; Xie, Y.; Li, Y.; Zhao, X.; Jiang, X.; Chen, D. Molecules 2018, 23, 222.
[https://doi.org/10.3390/molecules23010222]