Indole Derivatives and a Diketopiperazine from Chromobacterium violaceum
Abstract
Three indole derivatives (1–3) and a diketopiperazine (4) were isolated from the ethyl acetate extract of Chromobacterium violaceum. Their structures were elucidated based on the analysis of NMR and HR-MS data and by comparing those in the previous literature. The antibacterial activities of the isolated compounds were evaluated against Gram-positive bacteria, including human pathogenic methicillin-resistant Staphylococcus aureus (MRSA), Lacticaseibacillus paracasei subsp. paracasei, and Brevibacterium epidermidis. Compound 1 exhibited moderate antibacterial activity against all the three strains with MIC values ranging from 8.58 to 34.3 μg/mL.
Keywords:
Chromobacterium violaceum, Indole derivatives, Violacein, Diketopiperazine, Antibacterial activityIntroduction
In the present century, with the increasing threat of antimicrobial resistance, there is an urgent need to develop new antimicrobials.1 The exploration of natural products from microorganisms remains a crucial strategy for addressing antibiotic resistance. Microbe-derived metabolites have played a fundamental role in antibiotic drug development.2 The diversity of their structures serves as a rich source of inspiration to identify novel scaffolds for the development of new classes of antibiotics with novel mechanisms of action.3
Chromobacterium violaceum isolated from water and soil in tropical and subtropical regions is a facultatively anaerobic, Gram-negative, non-sporing, motile, catalase-positive, oxidase-positive, and rod-shaped bacteria.4 They are found to be a sessile bacterium in their natural habitat. As sessile bacteria are more prone to predation, their secondary metabolites could work as natural defense mechanisms, thereby giving them a competitive advantage in the habitat.5 Although C. violaceum is recognized for its ability to produce potent antibiotics like violacein, much of the research has centered on violacein itself.6–8 Therefore, with limited exploration into new antibiotics produced by this strain, our study aims to isolate and verify the antibiotic efficacy of these compounds.
Experimental
General experimental procedures – HR-ESI-MS data was obtained on an Agilent 6545 LC/QTOF spectrometer coupled to an Agilent Infinity II 1290 high-performance liquid chromatography (Agilent Technologies, Santa Clara, CA, USA) with a ZORBAX RRHD Eclipse Plus C18, 95 Å (2.1 mm × 50 mm i.d., 1.8 μm, Agilent Technologies, Santa Clara, CA, USA). 1H-NMR spectra were recorded on a Bruker AVANCE III 700 NMR spectrometer (Varian UNITY INOVA 500 spectrometer, USA). HPLC was conducted using an Agilent Infinity II 1260 High-performance liquid chromatography (HPLC) (Agilent Technologies, Santa Clara, CA, USA) with a column of Luna C18(2) 100 Å (250 mm × 10.0 mm i.d., 10 μm, Phenomenex, Torrance, CA, USA). Incubator IB3-03A (Jeiotech, Korea) and Shaking Incubator ISS-3075R (Jeiotech, Korea) were used for incubation of bacteria.
Bacterial sources – C. violaceum KCTC 2897 (or ATCC 12472), Brevibacterium epidermidis KCTC 3090, and Lacticaseibacillus paracasei subsp. paracasei KCTC 2897 were purchased from the Korean Collection for Type Cultures (KCTC), in August 2020. The frozen stocks of the bacteria were deposited in the −80˚C freezer of the School of Pharmacy, Sungkyunkwan University, Suwon, Korea. Methicillin-resistant Staphylococcus aureus (MRSA) USA300 was provided by Prof. Wonsik Lee, School of Pharmacy, Sungkyunkwan University.
Cultivation and extraction – After taking out the C. violaceum frozen stock stored in the −80˚C freezer, it was streaked on Luria-Bertani (LB) agar using a wood stick and incubated for overnight at 37°C. A single colony was collected from the LB agar plate, inoculated using a wood stick to 5 mL of LB broth in the polystyrene tube, and incubated for overnight in a shaking incubator at 37°C and 250 rpm. C. violaceum culture was 1:200 diluted by putting 25 μL of the bacterial culture into six 14-mL polypropylene tubes containing 5 mL of LB broth and incubated for 2 days in a shaking incubator at 37°C and 250 rpm. C. violaceum culture in the six tubes were inoculated in each 4 L Erlenmeyer flasks with 1 L of LB broth. These cultures were incubated for 2 days in the shaking incubator at 37°C and 250 rpm, which were extracted with ethyl acetate (6 L × 2 times).
Isolation and purification of the compounds – The dried extract (589 mg) was fractionated by HPLC using a C18(2) column with a gradient solvent system (CH3CN-H2O: 10/90 to 100/0 with 0.01% trifluoroacetic acid over 30 mins; flow rate: 4 mL/min; fraction collection: 1 min time window) to afford 30 fractions. Violacein (1) was eluted from fraction 17. It was purified with a C18(2) column and eluted at a retention time of 18.1 min (1.4 mg) under gradient solvent conditions (CH3CN-H2O: 30/70 to 40/60 with 0.01% trifluoroacetic acid over 30 min; flow rate: 4 mL/min). Deoxyviolacein and 1-acetyl-β-carboline (2 and 3) were eluted from the fraction 20. Deoxyviolacein (2) was purified (CH3CN-H2O: 20/80 to 50/50 with 0.01% trifluoroacetic acid over 30 min; flow rate: 4 mL/min) with an identical column and was obtained in its pure form after 25.5 min (0.8 mg). 1-Acetyl-β-carboline (3) was purified with a C18(2) column and eluted at a retention time of 22.5 min (0.8 mg) under gradient solvent conditions (CH3CN-H2O: 30/70 to 40/60 with 0.01% trifluoroacetic acid over 30 min; flow rate: 4 mL/min). Cyclo(L-Phe-L-Phe) (4) was eluted from the fraction 5. It was also purified (CH3CN-H2O: 25/75 to 35/65 with 0.01% trifluoroacetic acid over 30 min; flow rate: 4 mL/min) with an identical column and was obtained in its pure form at 15.2 min (0.7 mg).
Violacein (1) − Purplish gum; HR-ESI-MS (Positive-ion mode) m/z 344.1032 [M+H]+ (calcd for C20H14N3O3, 344.1030); 1H-NMR (DMSO-d6, 700 MHz): δ 11.88 (1H, s, NH-1), 10.71 (1H, s, NH-10), 10.60 (1H, s, NH-15), 9.32 (1H, s, OH-6), 8.93 (1H, d, J = 7.6 Hz, H-19), 8.07 (1H, s, H-2), 7.55 (1H, s, H-13), 7.34 (1H, d, J = 8.6 Hz, H-8), 7.23 (1H, s, H-5), 7.20 (1H, td, J = 7.6, 1.2 Hz, H-21), 6.95 (1H, td, J = 7.6, 1.2 Hz, H-20), 6.82 (1H, d, J = 7.6 Hz, H-22), 6.79 (1H, dd, J = 8.6, 2.3 Hz, H-7).
Deoxyviolacein (2) − Purplish gum; HR-ESI-MS (Positive-ion mode) m/z 328.1070 [M+H]+ (calcd for C20H14N3O2, 328.1081); 1H-NMR (CD3OD, 700 MHz): δ 8.99 (1H, d, J = 7.6 Hz, H-19), 8.01 (1H, dd, J = 6.7, 2.0 Hz, H-5), 7.94 (1H, s, H-2), 7.71 (1H, s, H-13), 7.51 (1H, dd, J = 6.7, 2.0 Hz, H-8), 7.29 (2H, m, H-6 and H-7), 7.22 (1H, td, J = 7.6, 1.2 Hz, H-21), 6.99 (1H, td, J = 7.6, 1.2 Hz, H-20), 6.87 (1H, d, J = 7.6 Hz, H-22).
1-Acetyl-β-carboline (3) − Colorless gum; HR-ESI-MS (Positive-ion mode) m/z 211.0860 [M+H]+ (calcd for C13H10N2O, 211.0866); 1H-NMR (CD3OD, 700 MHz): δ 8.47 (1H, d, J = 4.9 Hz, H-3), 8.32 (1H, d, J = 4.9 Hz, H-4), 8.23 (1H, dd, J = 7.8, 1.0 Hz, H-7), 7.71 (1H, d, J = 8.2 Hz, H-10), 7.60 (1H, ddd, J = 8.2, 7.0, 1.2 Hz, H-9), 7.32 (1H, ddd, J = 8.0, 7.0, 0.9 Hz, H-8), 2.83 (3H, s, H3-15).
Cyclo(L-Phe-L-Phe) (4) − Colorless gum; HR-ESI-MS (Positive-ion mode) m/z 295.1444 [M+H]+ (calcd for C18H18N2O2, 295.1441); 1H-NMR (CD3OD, 700 MHz): δ 7.34 (4H, tt, J = 7.9, 1.2 Hz, H-5, H-5′, H-7 and H-7′), 7.26 (2H, m, H-6 and H-6′), 7.11 (4H, dd, J = 7.9, 1.2 Hz, H-4, H-4′, H-8 and H-8′), 4.09 (2H, dd, J = 7.0, 4.1 Hz, H-1 and H-1′), 2.79 (2H, dd, J = 13.7, 4.1 Hz, H-2a and H-2′a), 2.15 (2H, dd, J = 13.7, 7.0 Hz, H-2b and H-2′b).
Antibacterial test – The antibacterial test was performed similarly to previous research.9–12 The three bacterial strains of MRSA, L. paracasei subsp. paracasei, and B. epidermidis were individually streaked on LB agar and incubated for 2 days at 37°C. A single colony was collected from each LB agar plate and inoculated into 5 mL of LB broth in a 14-mL polypropylene tube. It was incubated overnight in a shaking incubator at 37°C, 250 rpm. Then, the OD600 value of bacterial culture was adjusted to 0.001. A 196 μL of fresh LB medium and 4 μL of each compound stock solution (10 mM) dissolved in DMSO were added to the first column of a 96-well plate. Using a multichannel pipet, the solution was serially diluted from the second to the last column of the 96-well plate. Then, 100 μL of the bacterial culture mentioned above (OD600 = 0.001) was added to each well. Each experiment was performed three times. The plate was incubated at 37°C in a standing incubator overnight, and the minimum inhibitory concentration (MIC) value of each compound was acquired from the well where the bacteria were not alive as determined visually.
Results and Discussion
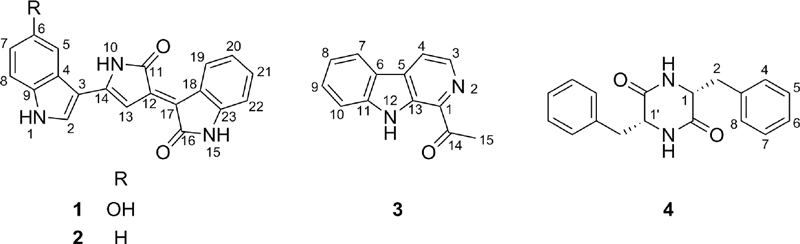
Three indole derivatives (1–3) along with a diketopiperazine (4) were isolated from the EtOAc extract of C. violaceum culture. The chemical structures of compounds 1−4 were determined as violacein (1),13,14 deoxyviolacein (2),13,14 1-acetyl-β-carboline (3),15 and cyclo(L-Phe-L-Phe) (4)16 (Fig. 1) by comparing their NMR and HR-MS data with those reported in the literatures.
Violacein (1) was obtained as a purplish gum. It showed UV-vis absorption peaks at 262, 375, and 573 nm, indicating the presence of a conjugated indole skeleton. Its molecular formula was determined to be C20H13N3O3 from the molecular ion peak [M+H]+ at m/z 344.1032 (calcd for C20H14N3O3, 344.1030) in the positive-ion HR-ESI-MS. The 1H-NMR spectrum of 1 displayed signals for three NH protons at δH 11.88 (1H, s, NH-1), 10.71 (1H, s, NH-10), and 10.60 (1H, s, NH-15), an OH proton at δH 9.32 (1H, s, OH-6), and nine aromatic protons at δH 8.93 (1H, d, J = 7.6 Hz, H-19), 8.07 (1H, s, H-2), 7.55 (1H, s, H-13), 7.34 (1H, d, J = 8.6 Hz, H-8), 7.23 (1H, s, H-5), 7.20 (1H, td, J = 7.6, 1.2 Hz, H-21), 6.95 (1H, td, J = 7.6, 1.2 Hz, H-20), 6.82 (1H, d, J = 7.6 Hz, H-22), and 6.79 (1H, dd, J = 8.6, 2.3 Hz, H-7). Based on these spectroscopic and spectrometric data analysis and the comparison with literature data, the structure of 1 was determined as violacein.13,14
The 1H-NMR and UV spectra of deoxyviolacein (2) closely resembled those of violacein (1). The difference of 1H-NMR in 2 was the presence of peaks for a 1,2-disubstituted benzene ring in 2 [δH 8.01 (1H, dd, J = 6.7, 2.0 Hz, H-5), 7.51 (1H, dd, J = 6.7, 2.0 Hz, H-8), and 7.29 (2H, m, H-6 and H-7)] instead of the 1,3,4-trisubstituted benzene ring in 1. HR-ESI-MS data of 2 indicated the molecular formula of 2 as C20H13N3O2, OH less than 1 and subsequently absence of the hydroxyl group at C-6 in 1. Collectively, the structure of 2 was characterized as deoxyviolacein by comparing its spectroscopic data with the literature values.13,14
1-Acetyl-β-carboline (3) was isolated as a colorless gum. Its molecular formula was established to be C13H10N2O based on positive-ion HR-ESI-MS, detected at m/z 211.0860 (cald for C13H10N2O, 211.0866). The 1H-NMR spectrum of 3 exhibited characteristic β-carboline signals [δH 8.47 (1H, d, J = 4.9 Hz, H-3), 8.32 (1H, d, J = 4.9 Hz, H-4), 8.23 (1H, dd, J = 7.8, 1.0 Hz, H-7), 7.71 (1H, d, J = 8.2 Hz, H-10), 7.60 (1H, ddd, J = 8.2, 7.0, 1.2 Hz, H-9), and 7.32 (1H, ddd, J = 8.0, 7.0, 0.9 Hz, H-8)] along with a single methyl signal at δH 2.83 (3H, s, H3-15) resulting from the substituted acetyl group at C-1. The chemical structure of compound 3 was determined by comparing 1H-NMR and HR-ESI-MS spectral data with those in the literature to be 1-acetyl-β-carboline.15
Cyclo(L-Phe-L-Phe) (4) was purified as a colorless gum. Its molecular formula was confirmed to be C18H18N2O2 from the molecular ion peak [M+H]+ at m/z 295.1444 (calcd for C18H18N2O2, 295.1444) in the positive-ion HR-ESI-MS. Compound 4 was identified through analysis of its 1H-NMR signals, including symmetric methine protons δH 4.09 (2H, dd, J = 7.0, 4.1 Hz, H-1 and H-1′) and methylene protons δH 2.79 (2H, dd, J = 13.7, 4.1 Hz, H-2a and H-2′a) and δH 2.15 (2H, dd, J = 13.7, 7.0 Hz, H-2b and H-2′b), along with signals from monosubstituted benzene protons [δH 7.34 (4H, tt, J = 7.9, 1.2 Hz, H-5, H-5′, H-7 and H-7′), 7.26 (2H, m, H-6 and H-6′), and 7.11 (4H, dd, J = 7.9, 1.2 Hz, H-4, H-4′, H-8 and H-8′)]. The chemical structure of compound 4 was established as cyclic diphenylalanine by analyzing its 1H-NMR and HR-ESI-MS spectral data and comparing them with published data.16
All the isolated compounds (1–4) were evaluated for their antibacterial activities against three Gram-positive bacteria including human pathogenic MRSA,17–20 L. paracasei subsp. paracasei, and B. epidermidis. Compound 1 exhibited moderate antibacterial activities against all three bacteria with MIC values of 8.6 μg/mL ( = 25 μM) for MRSA, 34.3 μg/mL ( = 100 μM) for L. paracasei subsp. paracasei, and 17.2 μg/mL ( = 50 μM) for B. epidermidis (Table 1). Compounds 2–4 were inactive against the tested bacteria (MIC > 100 μM). From these antibacterial activity data, we observed an interesting structure-activity relationship (SAR). The only structural difference between 1 and 2 is presence of a hydroxy group at C-6 in 1 which suggested that this functionality would play an important role in displaying antibacterial activity against Gram-positive bacteria. These antibacterial test results of violacein (1) are consistent with previous research.21
In this study, we report isolation and structure characterization of four secondary metabolites (1–4) from C. violaceum culture and their antibacterial activity against three Gram-positive bacteria, including a human pathogenic bacterium MRSA. Violacein (1) showed moderate antibacterial activity against all three tested bacteria, MRSA, L. paracasei subsp. paracasei, and B. epidermidis, with MIC values of 8.6–34.3 μg/mL. Also, we found that the hydroxy group at C-6 in 1 is essential for its antibacterial activity.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korean government (MSIT) (Nos. 2021R1C1C1011045 and 2022R1A6A1A03054419), by the BK21 FOUR Project.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
-
Miethke, M.; Pieroni, M.; Weber, T.; Brönstrup, M.; Hammann, P.; Halby, L.; Arimondo, P. B.; Glaser, P.; Aigle, B.; Bode, H. B.; Moreira, R.; Li, Y.; Luzhetskyy, A.; Medema, M. H.; Oernodet, J.-L.; Stadler, M.; Tormo, J. R.; Geniloud, O.; Truman, A. W.; Weissman, K. J.; Takano, E.; Sabatini, S.; Stegmann, M.; Empting, M.; Hirsch, A. K.; Loretz, B,; Legr, C.-M.; Titz, A.; Herrmann, J.; Jaeger, T.; Alt, S.; Hesterkamp, T.; Winterhalter, M.; Schiefer, A.; Pfarr, K.; Hoerauf, A.; Graz, H.; Graz, M.; Lindvall, M.; Ramurthy, S.; Karlén, A.; van Dongen, M.; Petkovic, H.; Keller, A.; Peyrane, F.; Donadio, S.; Fraisse, L.; Piddock, L. J. V.; Gilbert, I. H.; Moser, H. E.; Müller, R. Nat. Rev. Chem. 2021, 5, 726–749.
[https://doi.org/10.1038/s41570-021-00313-1]

-
Jiang, R.; Um, S.; Jeong, H.; Seo, J.; Huh, M.; Kim, Y. R.; Moon, K. Nat. Prod. Sci. 2023, 29, 59–66.
[https://doi.org/10.20307/nps.2023.29.2.59]

-
Schneider, Y. K. Antibiotics (Basel) 2021, 10, 842.
[https://doi.org/10.3390/antibiotics10070842]

- Richard, C. Bull. Soc. Pathol. Exot. 1993, 86, 169–173.
-
Choi, S. Y.; Yoon, K.-H.; Lee, J. I.; Mitchell, R. J. Biomed Res. Int. 2015, 2015, 465056.
[https://doi.org/10.1155/2015/465056]

-
Konzen, M.; De Marco, D.; Cordova, C. A.; Vieira, T. O.; Antônio, R. V.; Creczynski-Pasa, T. B. Bioorg. Med. Chem. 2006, 14, 8307–8313.
[https://doi.org/10.1016/j.bmc.2006.09.013]

-
De Carvalho, D. D.; Costa, F. T. M.; Duran, N.; Haun, M. Toxicol. In Vitro 2006, 20, 1514–1521.
[https://doi.org/10.1016/j.tiv.2006.06.007]

-
Masuelli, L.; Pantanella, F.; La Regina, G.; Benvenuto, M.; Fantini, M.; Mattera, R.; Di Stefano, E.; Mattei, M.; Silvestri, R.; Schippa, S. Tumor Biol. 2016, 37, 3705–3717.
[https://doi.org/10.1007/s13277-015-4207-3]

-
Ham, S. L.; Lee, T. H.; Kim, K. J.; Kim, J. H.; Hwang, S. J.; Lee, S. H.; Yu, J. S.; Kim, K. H.; Lee, H.-J.; Lee, W.; Kim, C. S. J. Nat. Prod. 2023, 86, 850–859.
[https://doi.org/10.1021/acs.jnatprod.2c01032]

-
Kim, H. R.; Kim, J.; Yu, J. S.; Lee, B. S.; Kim, K. H.; Kim, C. S. Arch. Pharm. Res. 2023, 46, 35–43.
[https://doi.org/10.1007/s12272-022-01424-z]

-
Lee, D. Y.; Kim, J.; Lee, G. S.; Park, S.; Song, J.; Lee, B. S.; Lee, S. R.; Kim, K. H.; Kim, C. S. ACS Chem. Biol. 2024, 19, 973–980.
[https://doi.org/10.1021/acschembio.3c00798]

-
Benaissa, A.; Abdelmounaim, K.; Benbelaid, F.; Bousselham, A.; Benziane, Y.; Muselli, A.; Bendahou, M. Nat. Prod. Sci. 2023, 29, 225–234.
[https://doi.org/10.20307/nps.2023.29.4.225]

-
Hoshino, T.; Kondo, T.; Uchiyama, T.; Ogasawara, N. Agric. Biol. Chem. 1987, 51, 965–968.
[https://doi.org/10.1080/00021369.1987.10868084]

-
Matz, C.; Webb, J. S.; Schupp, P. J.; Phang, S. Y.; Penesyan, A.; Egan, S.; Steinberg, P.; Kjelleberg, S. PLoS One 2008, 3, e2744.
[https://doi.org/10.1371/journal.pone.0002744]

-
Zhou, T.-S.; Ye, W.-C.; Wang, Z.-T.; Che, C.-T.; Zhou, R.-H.; Xu, G.-J.; Xu, L.-S. Phytochemistry 1998, 49, 1807–1809.
[https://doi.org/10.1016/S0031-9422(98)00232-5]

-
Wang, J.-M.; Ding, G.-Z.; Fang, L.; Dai, J.-G.; Yu, S.-S.; Wang, Y.- H.; Chen, X.-G.; Ma, S.-G.; Qu, J.; Xu, S.; Du, A.; J. Nat. Prod. 2010, 73, 1240–1249.
[https://doi.org/10.1021/np1000895]

- Kang, D. H.; Kang, O. H.; Chae, H.-S.; Kwon, D. Y. Kor. J. Pharmacogn. 2023, 54, 72–79.
-
Kwon, Y. Kor. J. Pharmacogn. 2023, 54, 145–152.
[https://doi.org/10.22889/KJP.2023.54.4.145]

-
Wei, Y.-M.; Tan, J.-S.; Tang, H.-W.; Tong, W.-Y.; Leong, C.-R.; Tan, W.-N. Nat. Prod. Sci. 2022, 28, 93–104.
[https://doi.org/10.20307/nps.2022.28.3.93]

- Hwang, H.; Kang, O.-H.; Kwon, D.-Y. Kor. J. Pharmacogn. 2022, 53, 87–95.
-
Aruldass, C. A.; Masalamany, S. R. L.; Venil, C. K.; Ahmad, W. A. Environ. Sci. Pollut. Res. 2018, 25, 5164–5180.
[https://doi.org/10.1007/s11356-017-8855-2]