Phenolic Compounds Isolated from Juncus decipiens and Their Effects on Osteoblast Differentiation in the Mouse Mesenchymal Stem Cell Line C3H10T1/2
Abstract
As part of our current projects to discover biologically active compounds from natural sources, we conducted a phytochemical investigation of Juncus decipiens, a species generally distributed throughout Korea. J. decipiens has been used in traditional Chinese medicine to control diuresis for strangury and clear out heart fire. The phytochemical investigation of the EtOH extract of J. decipiens led to the isolation of five phenolic compounds (1–5) via semi-preparative HPLC purification. The chemical structures of the isolated compounds were identified as isosaponarin (1), isovitexin 7,2′′-di-O-glucoside (2), 4-O-feruloylquinic acid (3), 5-O-feruloylquinic acid (4), and 3-O-caffeoylquinic acid (5) based on comparisons of their spectroscopic and physical properties with those reported in previous studies. Notably, this is the first report of the presence of compounds (1–4) in J. decipiens. Then, compounds 1–5 were tested to determine their effects on osteogenesis and adipogenesis in the mouse mesenchymal stem cell line C3H10T1/2. We found that quinic acid derivatives (3–5) promoted the osteogenic differentiation of stem cells. These findings demonstrate that the bioactive quinic acid derivatives might be effective for the treatment of menopause-associated syndromes, such as osteoporosis, as the isolated compounds were shown to promote osteogenesis of stem cells.
Keywords:
Juncus decipiens, Juncaceae, Quinic acid derivatives, OsteogenesisIntroduction
As part of our continuous endeavor to discover structurally and biologically novel compounds from diverse natural sources,1–5 we have collected and prepared ethanol (EtOH) extracts of freshwater plants for phytochemical investigation. Among the collected freshwater plants, Juncus decipiens, a member of the Juncaceae family, possesses culinary, medicinal, and decorative properties. It is generally distributed throughout Korea and the temperate and subtropical regions of the northern hemisphere. For many years, J. decipiens has been used in traditional Chinese medicine to control diuresis for strangury and clear out heart fire.6 Previous studies on this plant have reported its pharmacological properties, including antithrombotic and hepatoprotective activities.7 It also exhibits anti-inflammatory activities by suppressing the production of inflammatory mediators in LPS-stimulated RAW 264.7 cells and by attenuating edema in mice.8
Recently, J. decipiens has gained medicinal attention as a source of phenanthrenes, phenolic compounds, glycerides, and flavonoids.9–12 One of the most interesting molecules identified in J. decipiens is phenanthrene-type compounds, which have shown potential in preventing Alzheimer’s disease13 and exhibiting antimicrobial activity.14 Its dimers have demonstrated bioactivities such as cytotoxicity and anti-inflammatory effects.15 Despite several previous reports, there have been few studies describing the biologically active phenolic constituents from J. decipiens.
In this study, we performed an extensive phytochemical analysis of the whole plant of J. decipiens. The column chromatographic separation of its EtOH extract and subsequent high-performance liquid chromatography (HPLC) purification led to the isolation of five phenolic compounds (1–5) via liquid chromatography/mass spectrometry (LC/MS)-based analysis. Their structures were elucidated by combining spectroscopic data and LC/MS analysis results. The isolated compounds 1–5 were then tested to determine their effects on osteogenesis in the mouse mesenchymal stem cell (MSC) line C3H10T1/2. Herein, we describe the isolation and structural characterization of the five phenolic compounds (1–5) and their potential effects on the reciprocal regulation of osteoblast differentiation.
Experimental
General experimental procedures – The optical rotations were obtained using a Jasco P-1020 polarimeter (Jasco, Easton, MD, USA). The nuclear magnetic resonance (NMR) spectra were obtained using a Bruker AVANCE III HD 850 NMR spectrometer at 850 MHz (1H) and 212.5 MHz (13C), with chemical shifts given in ppm (δ). Preparative HPLC was performed using a Waters 1525 Binary HPLC pump with a Waters 996 Photodiode Array Detector (Waters Corporation, Milford, CT, USA). Semi-preparative HPLC was conducted using a Waters HPLC System with 2998 photodiode array detectors (Waters Corporation, Milford, CT, USA) and Waters 2690 binary HPLC pump. LC/MS analysis was carried out on an Agilent 1200 Series HPLC system (Agilent Technologies, Santa Clara, CA, USA) equipped with a diode array detector and a 6130 Series ESI mass spectrometer by using an analytical Kinetex C18 100 Å column (100 mm × 2.1 mm i.d., 5 μm) (Phenomenex, Torrance, CA). High-resolution (HR) electrospray ionization (ESI) mass spectra were recorded on an Agilent 6545 QTOF LC/MS spectrometer (Agilent Technologies, Santa Clara, CA, USA). Merck precoated silica gel F254 plates and RP-18 F254s plates were used for thin-layer chromatography (TLC). Spots were detected on TLC under UV light or by heating after spraying with anisaldehyde-sulfuric acid.
Plant material – The whole plant of J. decipiens (Buchenau) Nakai were collected at Jecheon-si, Chungcheongbuk-do, Replublic of Korea, in May 2023. The plant material was verified by Dr. Moon Jin Ra, Jeong Nam Yu, and Sang Mi Jung at Nakdonggang National Institute of Biological Resources, Sangju, Republic of Korea. A voucher specimen, namely HIMH-2304, was stored at the herbarium of the School of Pharmacy affiliated in Sungkyunkwan University, Suwon, Republic of Korea.
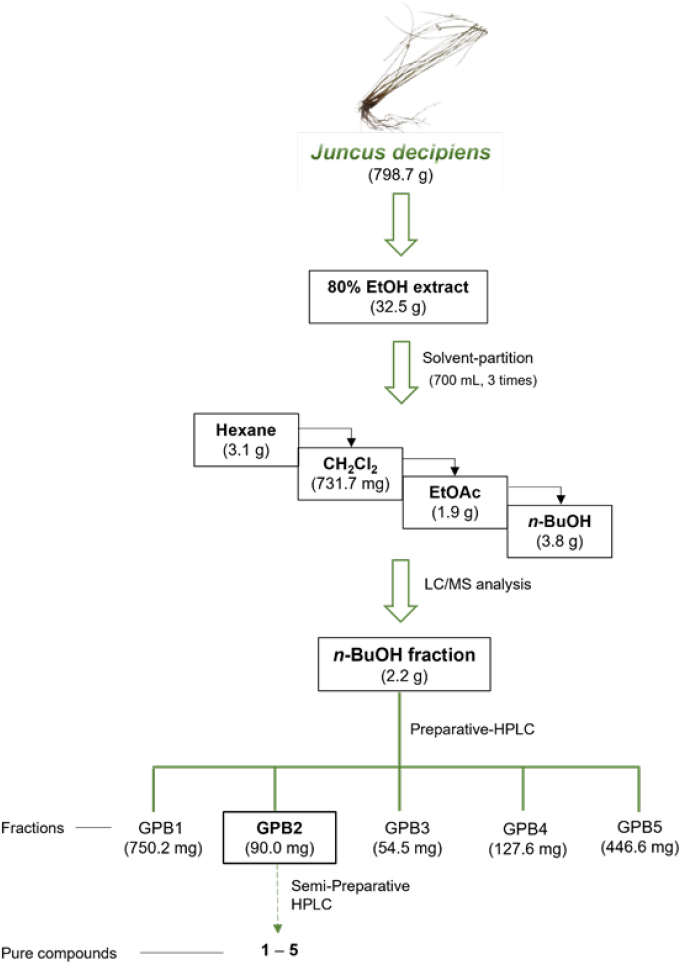
Extraction and isolation – Completely dried whole plants of J. decipiens (798.7 g) were extracted by sonicating three times (2.0 L×3) for 90 min in 80% ethanol (EtOH) at room temperature and then filtered. The filtrate was subsequently evaporated in vacuo to obtain a crude EtOH extract (35.2 g). This extract was dissolved in distilled water (700 mL) and then solvent-partitioned with n-hexane, dichloromethane (CH₂Cl₂), ethyl acetate (EtOAc), and n-butanol (n-BuOH) (each 700 mL×3), yielding 3.1 g, 731.7 mg, 1.9 g, and 3.8 g of extract, respectively. Through LC/MS analysis of each fraction, we found that the n-BuOH-soluble fraction harbors the main phenolic compounds with characteristic UV patterns of flavonoids. Consequently, 2.2 g of the n-BuOH fraction (3.8 g) was collected and divided into five fractions (GPB1-GPB5) using preparative HPLC. Five subfractions were obtained from the n-BuOH fraction (2.2 g) by preparative reversed-phase HPLC (Phenomenex Luna C18, 250×21.2 mm i.d., 5 μm) using an isocratic system of 37% MeOH/H₂O at a flow rate of 5 mL/min. From subfraction GPB2 (90.0 mg), compounds 1 (2.6 mg, tR = 55.0 min), 2 (2.6 mg, tR = 57.0 min), 3 (0.6 mg, tR = 63.0 min), and 4 (7.5 mg, tR = 74.0 min), 5 (43.4 mg, tR = 38.0 min) were isolated via semi-preparative reversed-phase HPLC (Phenomenex C18 100 Å column, 250×2.1 mm i.d., 5 μm) using a gradient system of 15–35% MeOH/H₂O at a flow rate of 2 mL/min.
Isosaponarin (1) – Yellow amorphous solid; : −29° (c 0.13, pyridine), Electrospray ionization-mass spectrometry (ESI-MS) m/z 595.2 [M+H]+; 1H-NMR (DMSO-d6, 850 MHz): δ 8.02 (2H, d, 9.0, J = H-2′ and H-6′), 7.18 (2H, d, 9.0, J = H-3′ and H-5’), 6.84 (1H, s, H-3), 6.52 (1H, s, H-8), Glc: 5.02 (1H, d, J = 7.5 Hz, H-1′′), 4.58 (1H, d, J = 10.0 Hz, H-1′′′), 4.07 (1H, t, J = 9.0 Hz, H-2′′′), 3.69 (1H, dd, 12.0, 2.0 Hz , H-6′′), 3.67 (1H, d, J = 11.0 Hz, H-6′′′), 3.48 (1H, dd, J = 12.0, 5.5 Hz, H-6′′), 3.29–3.32 (2H, m, overlapped, H-3′′ and H-5′′), 3.25–3.29 (1H, m, overlapped, H-2′′), 3.19 (1H, dd, J = 18.0, 9.0 Hz, H-5′′′), 3.17 (2H, m, H-3′′′ and H-4’’); 13C-NMR (DMSO-d6, 212.5 MHz): δ 181.7 (C-4), 162.5 (C-7), 162.4 (C-2), 160.7 (C-5), 160.2 (C-4′), 156.4 (C-9), 128.1 (C-6′), 124.0 (C-1′), 116.6 (C-3′ and C-5’), 109.1 (C-6), 103.7 (C-3), 99.9 (C-1′′), 94.0 (C-8), 81.5 (C-5′′′), 79.1 (C-3′′′), 77.2 (C-5′′), 76.5 (C-3′′), 73.2 (C-2′′ and C-1′′′), 70.6 (C-4′′′), 70.1 (C-2′′′), 69.6 (C-4′′), 61.5 (C-6′′′), 60.6 (C-6′′).
Isovitexin 7,2′′-di-O-glucoside (2) – Yellow amorphous powder; : −62.0° (c 0.13, MeOH), ESI-MS m/z 757.2 [M+H]+; 1H-NMR (CD3OD, 850 MHz): δ 8.00 (2H, d, J = 8.5 Hz, H-2′ and H-6′), 7.27 (2H, d, J = 8.5 Hz, H-3′ and H-5′), 6.73 (1H, s, H-3), 6.60 (1H, s, H-6), Glc: 5.16 (1H, d, J = 7.5 Hz, H-1′′), 4.90 (1H, d, J = 9.5 Hz, H-1′′′), 4.50 (1H, d, J = 7.5 Hz, H-1′′′′), 3.38–3.77 (18H, m, overlapped, H-2′′–H-6′′, H-2′′′–H-6′′′, and H-2′′′′–H-6′′′′).
4-O-feruloylquinic acid (3) – White powder; : −55.7° (c 0.03, MeOH), ESI-MS m/z 369.1 [M+H]+; 1H-NMR (CD3OD+D2O, 850 MHz): δ 7.65 (1H, d, J = 16.0 Hz, H-7′), 7.14 (1H, d, J = 2.0 Hz, H-2′), 7.04 (1H, dd, J = 8.5, 2.0 Hz, H-6′), 6.85 (1H, d, J = 8.5 Hz, H-5′), 6.42 (1H, d, J = 16.0, H-8′), 4.85 (1H, dd, J = 9.0, 3.0 Hz, H-4), 4.31 (1H, dt, J = 6.5, 3.0 Hz, H-3), 4.27 (1H, td, J = 9.0, 4.0 Hz, H-5), 3.77 (3H, s, OCH3), 2.22 (1H, dd, J = 14.0, 3.0 Hz, H-2a), 2.19 (1H, m, H-6a), 2.07 (1H, m, H-2a), 2.06 (1H, m, H-6b).
5-O-feruloylquinic acid (4) – White powder; : +10.9° (c 0.38, MeOH), ESI-MS m/z 369.1 [M+H]+; 1H-NMR (CD3OD+D2O, 850 MHz): δ 7.53 (1H, d, J = 16.0 Hz, H-7′), 7.08 (1H, d, J = 2.0 Hz, H-2′), 6.97 (1H, dd, J = 8.5, 2.0 Hz, H-6′), 6.82 (1H, d, J = 8.5 Hz, H-5′), 6.24 (1H, d, J = 16.0, H-8′), 5.27 (1H, dd, J = 12.0, 7.0 Hz, H-4), 4.16 (1H, dt, J = 7.0, 3.0 Hz, H-4), 3.77 (1H, dd, J = 7.5, 3.0 Hz, H-3), 3.70 (3H, s, OCH3), 2.22 (1H, dd, J = 14.0, 3.0 Hz, H-6a), 2.18 (1H, m, H-2b), 2.06 (1H, m, H-2b), 2.02 (1H, m, H-6b).
3-O-caffeoylquinic acid (5) – Colorless needles; : −134.6° (c 2.17, MeOH), ESI-MS m/z 355.1 [M+H]+; 1H-NMR (CD3OD, 850 MHz): δ 7.56 (1H, d, J = 16.0 Hz, H-7′), 7.05 (1H, d, J = 2.0 Hz, H-2′), 6.95 (1H, dd, J = 8.0, 2.0 Hz, H-6′), 6.78 (1H, d, J = 8.0 Hz, H-5′), 5.34 (1H, dd, J = 12.0, 8.5 Hz, H-3), 4.18 (1H, s, H-5), 3.73 (1H, dd, J = 8.5, 3.0 Hz, H-4), 2.16–2.22 (2H, m, H-2a and H-6a), 2.15–2.02 (2H, m, H-2b and H-6b).
LC-MS analysis for fraction sand 3D plot visualization – The LC-MS analysis was carried out using an Agilent 1200 series HPLC system, equipped with a diode array detector and a 6130 Series ESI mass spectrometer. For the analysis, an analytical Kinetex C18 100 Å column (100 × 2.1 mm i.d., 5 μm particle size) from Phenomenex (Torrance, CA, USA), was employed, with a flow rate maintained at 0.3 mL/min. The fraction samples were analyzed using a gradient elution program from 10% MeOH-H2O to 100% MeOH for 52 min [10% MeOH-H2O → 100% MeOH (0–30 min), 100% MeOH (30–41 min), 100% MeOH → 10% MeOH-H2O (41–42 min), 10% MeOH-H2O (42–52 min), and a flow rate of 0.3 mL/min]. For the LC-MS data analysis, the application MZmine 4.1.0 was used to process the 3D plot visualization of the LC/MS data. The parameters were set as follows: MS mass range 100-1000 Da, retention time resolution 500, m/z resolution 500, MS level 1, Polarity +, Spectrum type Centroided.
Cell culture – The C3H10T1/2 cell line, which originated from mouse embryonic fibroblasts, was cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 10% heat-inactivated fetal bovine serum, 100 U/mL of penicillin, and 100 μg/mL of streptomycin at 37°C with 5% CO2. To measure adipogenic activity, C3H10T1/2 cells were plated in a six-well plate at a density of 5 × 105 cells/mL. The cells were then treated with 1 μM of dexamethasone (DMS), 10 μM of troglitazone, 5 μg/mL of insulin, and 0.5 mM of 3-isobutyl-1-methylxanthine for 48 h. Subsequently, the cells were cultured for an additional 72 h with 10 μM of troglitazone and 5 μg/mL of insulin. To measure osteoblastic activity, C3H10T1/2 cells were plated in a six-well plate at a density of 5 × 105 cells and treated with 10 mM of β-glycerophosphate and 50 μg/mL of ascorbic acid. The culture medium was changed every 72 h.
Oil Red O staining – The cultured cells were rinsed with phosphate-buffered saline (PBS) and fixed in 10% neutral-buffered formalin at room temperature for 1 hour. Subsequently, the cells were stained with a 0.5% filtered stock solution of Oil Red O in isopropanol (Sigma, Saint Louis, MO, USA). After staining, the intracellular triglyceride content was assessed by redissolving the stained cells in isopropanol, followed by measurement of absorbance at a wavelength of 520 nm.
Alkaline phosphatase staining – Cultured cells were washed with 2 mM of MgCl2 and then incubated with an alkaline phosphatase (ALP) buffer (100 mM of Tris-HCl, pH 9.5; 100 mM of NaCl; 10 mM of MgCl2; and 0.05% Tween-20) for 15 min. The cells were then incubated in an ALP buffer containing 0.4 mg/mL of nitro-blue tetrazolium (Sigma) and 0.2 g/mL of 5-bromo-4-chloro-3-indolyl phosphate (Sigma). After washing with 0.5 mM of ethylenediaminetetraacetic acid (EDTA), the cells were fixed with 10% neutral-buffered formalin for 1 h.
mRNA isolation and real-time polymerase chain reaction (PCR) – RNA extraction from the cells was performed using the NucleoZOL reagent (NucleoZOL; Macherey-Nagel GmbH & Co., KG, Germany). Subsequently, complementary DNA (cDNA) was synthesized from 0.5 μg of total RNA utilizing a ReverTraAce qPCR reverse transcription (RT) Master Mix Kit (FSQ-201; Toyobo, Japan) with random primers. The resulting cDNA was then combined with the amplification mixture comprising the Thunderbird SYBR qPCR Mix (Toyobo) along with the specified primers. The primers used for the quantitative PCR were:
ALP: F (ATC GGA ACA ACC TGA CTG ACC) and R (ACC CTC ATG ATG TCC GTG GTC AAT).
Osteopontin: F (GAC CAC ATG GAC GAC GAT G) and R (TGG AAC TTG CTT GAC TAT CGA
Osterix: F (GAT GGC GTC CTC TCT GCT T) and R (CGT ATG GCT TCT TTG TGC CT).
Statistical analysis – One-way analysis of variance was used to determine whether there were any statistically significant differences between the control group and the test group.
Results and Discussions
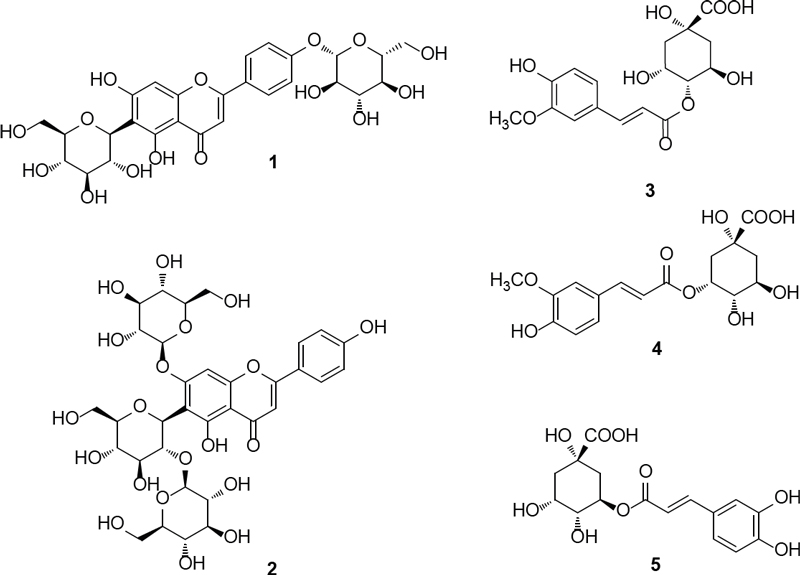
The EtOH extract of the whole plant of J. decipiens underwent solvent partitioning using four organic solvents: n-hexane, dichloromethane (CH₂Cl₂), ethyl acetate (EtOAc), and n-butanol (n-BuOH). This process afforded four main fractions: n-hexane, CH₂Cl₂, EtOAc, and n-BuOH soluble fractions (Fig. 1). Each fraction was analyzed by LC-MS in positive ion mode, and the 3D plot visualization of the LC/MS data was processed using the application MZmine 4.1.0. The 3D plot visualization showed that the n-BuOH-soluble fraction contained major peaks (Fig. 2), including phenolic compounds exhibiting a characteristic UV spectrum of flavonoids. The 3D plot visualization of subfractions from the n-BuOH-soluble fraction indicated that the subfraction GPB2 is a promising fraction for investigating the main phenolic compounds (Fig. 2). The chemical analysis of the subfraction GPB2 using semi-preparative HPLC resulted in the purification of five phenolic compounds (1–5) (Fig. 1) which include two flavonoid glycosides (1 and 2) and three quinic acid derivatives (3–5). The compounds were identified as isosaponarin (1),16 isovitexin 7,2′′-di-O-glucoside (2),17 4-O-feruloylquinic acid (3),18 5-O-feruloylquinic acid (4),18 and 3-O-caffeoylquinic acid (5)19 by comparing their spectroscopic data with reported values from previously published studies (Fig. 3). Notably, compound 1, isosaponarin, has been previously identified as a major substance in wasabi leaves and reported for its bioactivity in regulating glutamate release.20,21 Compound 2, isovitexin 7,2′′-di-O-glucoside, was first isolated and reported from Cerastium arvense.22 Compound 3, 4-O-feruloylquinic acid, and compound 4, 5-O-feruloylquinic acid, have been isolated from green coffee beans and are known for their inhibitory effects on porcine pancreas α-amylase isozyme I and they have been reported to possess antioxidant activity.23 Compound 5, 3-O-caffeoylquinic acid, has been primarily isolated from several plants and exhibits various bioactivities, including antioxidant, antitumor, anti-inflammatory, antibacterial, protective effects on blood vessels and the nervous system, and regulation of carbohydrate and lipid metabolism.24 Based on a literature search, to the best of our knowledge, this is the first report of the presence of four phenolic compounds (1–4) from J. decipiens.
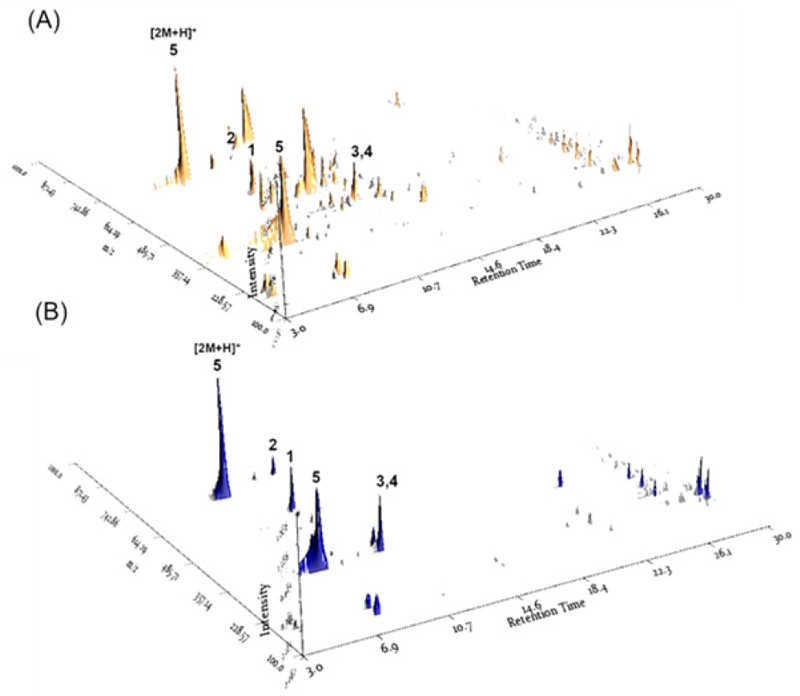
The 3D plot visualization of the LC/MS data from the n-BuOH-soluble fraction (A) and subfraction GPB2 from the n-BuOH-soluble fraction (B). The peak assignment for compounds 1–5 was based on the molecular ion peak of [M+H]+, except for an additional peak observed for compound 5.
Mesenchymal stem cells (MSCs) in the bone marrow are pluripotent cells capable of differentiating into osteocytes and adipocytes. Microenvironmental changes influence the regulation of gene expression during MSC differentiation, and alterations in the expression of these genes may disrupt the balance between osteoprogenitor and adipocyte progenitor cells in osteoporosis patients.25–27 Therefore, a therapy that can modulate gene expression in MSCs holds promise for managing postmenopausal osteoporosis.
To evaluate the therapeutic effects of compounds 1–5 on MSC differentiation, we investigated their influence on the differentiation of murine MSCs into adipocytes and osteoblasts. The murine mesenchymal stem cell line C3H10T1/2 was treated with 10 μM of each compound during adipogenesis. Differentiated cells were stained with Oil Red O (ORO) to assess lipid droplet formation. Compound 5 was observed to slightly reduce lipid droplet formation, resulting in fewer ORO-stained cells compared to normally differentiated adipocytes (Fig. 4A and 4B). In osteogenesis assays, MSCs were cultured in osteogenesis-inducing media in the presence of compounds 1–5 and stained to detect alkaline phosphatase (ALP) production, a marker of osteoblast differentiation.28 Compounds 3–5 induced a slight increase in ALP staining intensity and stimulated ALP enzyme activity compared to the negative control group (Fig. 4C and 4D).
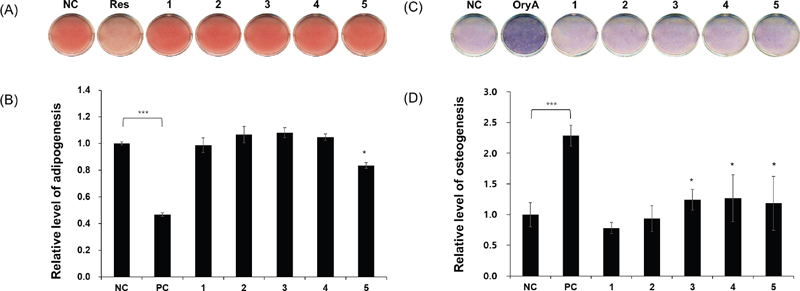
The effects of compounds 1–5 on the differentiation of MSCs into osteoblasts or adipocytes. The mouse MSC line C3H10T1/2 was treated with compounds 1–5. After adipogenic differentiation, the cells were stained with ORO (A) and the absorbance of the stained lipid droplets was quantified (B). Osteoblast differentiation was assessed using alkaline phosphatase (ALP) staining (C), with the intensity of the stained cells measured (D). The untreated negative control is labeled as NC. Adipogenesis was induced using 20 μM resveratrol (Res) as a positive control (PC), and osteogenesis was induced using 5 μM oryzativol A (OryA). Each compound was applied at a concentration of 10 μM in media inducing adipogenesis or osteogenesis. Statistical significance: *p < 0.1; ***p < 0.005.
Since compounds 3–5 were found to promote osteogenic differentiation of MSCs, we investigated whether these compounds affect the expression of osteogenesis-related genes during the differentiation process. Compound 3 significantly upregulated the gene expression of alkaline phosphatase (ALP) (Fig. 5). Additionally, compound 5 marginally increased the expression of osterix, a key transcription factor in osteogenesis, during osteogenic differentiation (Fig. 5). These findings indicate that compounds 3 and 5 not only enhance osteogenic differentiation at the cellular level but also modulate the expression of critical genes involved in osteoblast formation, further supporting their potential as therapeutic agents for osteoporosis and other bone-related disorders.
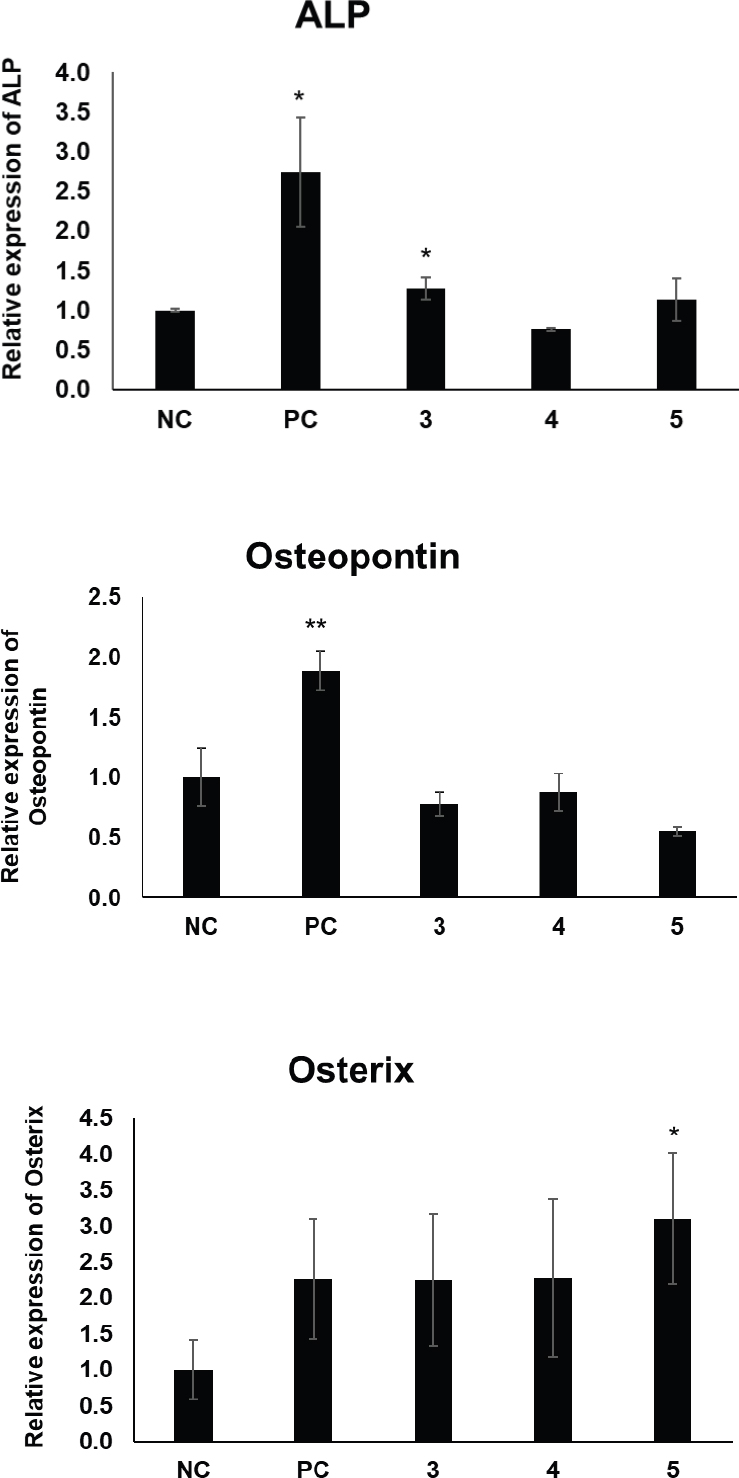
Gene expression levels of osteogenic markers ALP, Ostepontin, and osterix in MSCs treated with compounds 3–5. Oryzativol A (OryA, 5 μM) was used as a positive control. Statistical significance: *p ≤ 0.1; **p < 0.01.
In conclusion, we identified five phenolic compounds (1–5) present in the whole plants of J. decipiens via LC/MS-based analysis. The chemical structures of the isolated compounds were identified as isosaponarin (1), isovitexin 7,2′′-di-O-glucoside (2), 4-O-feruloylquinic acid (3), 5-O-feruloylquinic acid (4), and 3-O-caffeoylquinic acid (5) based on comparisons of their spectroscopic and physical properties with those reported in previous studies. To the best of our knowledge, this is the first report of the presence of compounds (1–4) in J. decipiens. We examined the effects of the isolated compounds on MSC differentiation into osteoblasts and found that three quinic acid derivatives (3–5) have the potential to modulate MSC differentiation towards osteogenesis over adipogenesis. Specifically, compound 3 significantly upregulated the expression of alkaline phosphatase (ALP), while compound 5 marginally increased the expression of osterix, a key factor in osteogenesis. Taken together, our results suggest that these bioactive compounds may be useful as an effective treatment for menopause-associated syndromes, such as osteoporosis.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (2019R1A5A2027340 and 2021R1A2C2007937). This work was also supported by the Korea Environment Industry & Technology Institute (KEITI) through a project to improve the development of multi-ministerial national biological research resources, funded by the Republic of Korea Ministry of Environment (MOE) (grant number 2021003420003).
Conflicts of Interest
The authors have no financial conflicts of interest to declare.
References
-
Lee, B. S.; So, H. M.; Kim, S.; Kim, J. K.; Kim, J.-C.; Kang, D.-M.; Ahn, M.-J.; Ko, Y.-J.; Kim, K. H. Arch. Pharm. Res. 2022, 45, 795–805.
[https://doi.org/10.1007/s12272-022-01416-z]

-
Cho, H.; Kim, K. H.; Han, S. H.; Kim, H.-J.; Cho, I.-H.; Lee, S. Nat. Prod. Sci. 2022, 28, 181–186.
[https://doi.org/10.20307/nps.2022.28.4.181]

-
Yu, J. S.; Jeong, S. Y.; Li, C.; Oh, T.; Kwon, M.; Ahn, J. S.; Ko, S.-K.; Ko, Y.-J.; Cao, S.; Kim, K. H. Arch. Pharm. Res. 2022, 45, 105–113.
[https://doi.org/10.1007/s12272-022-01372-8]

-
Lee, S. R.; Lee, B. S.; Yu, J. S.; Kang, H.; Yoo, M. J.; Yi, S. A.; Han, J.-W.; Kim, S.; Kim, J. K.; Kim, J.-C.; Kim, K. H. J. Ginseng Res. 2022, 46, 357–366.
[https://doi.org/10.1016/j.jgr.2021.09.004]

-
Lee, D. E.; Park, K. H.; Hong, J.-H.; Kim, S. H.; Park, K.-M.; Kim, K. H. Arch. Pharm. Res. 2023, 46, 771–781.
[https://doi.org/10.1007/s12272-023-01468-9]

-
Ullah, S.; Amen, Y.; Shimizu, K. Nat. Prod. Res. 2023, 1–11.
[https://doi.org/10.1080/14786419.2023.2223749]

-
Cheng, J.; Zhang, M.; Sun, Z.; Lu, F.; Xiong, W.; Luo, J.; Kong, H.; Wang, Q.; Qu, H.; Zhao, Y. Nanomedicine (Lond) 2019, 14, 431–446.
[https://doi.org/10.2217/nnm-2018-0285]

-
Park, N.-Y.; Kim, S.-G.; Park, H.-H.; Jeong, K.-T.; Lee, Y. J.; Lee, E. Pharm. Biol. 2016, 54, 243–250.
[https://doi.org/10.3109/13880209.2015.1029053]

-
Shima, K.; Toyota, M.; Asakawa, Y. Phytochemistry 1991, 30, 3149–3151.
[https://doi.org/10.1016/S0031-9422(00)98276-1]

- Kim, Y. J.; Park, C. I.; Park, J. S.; Ahn, E. M. J. Appl. Biol. Chem. 2014, 57, 47–51.
-
Ishiuchi, K.; Kosuge, Y.; Hamagami, H.; Ozaki, M.; Ishige, K.; Ito, Y.; Kitanaka, S. J. Nat. Med. 2015, 69, 421–426.
[https://doi.org/10.1007/s11418-015-0898-4]

- Chen, L.-C.; Huang, C.-M.; Chu, Y.-C.; Hsieh, Y.-H.; Cheng, M.-J.; Hsiao, J.-W.; Chen, J.-J. Chem. Nat. Compd. 2024, 1–5.
-
Tamano, H.; Takiguchi, M.; Saeki, N.; Katahira, M.; Shioya, A.; Tanaka, Y.; Egawa, M.; Fukuda, T.; Ikeda, H.; Takeda, A. Mol. Neurobiol. 2021, 58, 3603–3613.
[https://doi.org/10.1007/s12035-021-02364-3]

-
Zhao, W.; Xu, L.-L.; Zhang, X.; Gong, X.-W.; Zhu, D.-L.; Xu, X.-H.; Wang, F.; Yang, X.-L. Fitoterapia 2018, 130, 247–250.
[https://doi.org/10.1016/j.fitote.2018.09.007]

-
Ma, W.; Liu, F.; Ding, Y.-Y.; Zhang, Y.; Li, N. Fitoterapia 2015, 105, 83–88.
[https://doi.org/10.1016/j.fitote.2015.06.006]

-
Hosoya, T.; Yun, Y. S.; Kunugi, A. Tetrahedron 2005, 61, 7037–7044.
[https://doi.org/10.1016/j.tet.2005.04.061]

-
Chen, J.-M.; Wei, L.-B.; Lu, C.-L.; Zhou, G.-X. J. Asian Nat. Prod. Res. 2013, 15, 1088–1093.
[https://doi.org/10.1080/10286020.2013.814107]

-
Ao, X.; Yan, J.; Liu, S.; Chen, S.; Zou, L.; Yang, Y.; He, L.; Li, S.; Liu, A.; Zhao, K. Food Chem. 2022, 374, 131743.
[https://doi.org/10.1016/j.foodchem.2021.131743]

-
Choi, Y. H.; Kim, H. K.; Linthorst, H. J. M.; Hollander, J. G.; Lefeber, A. W. M.; Erkelens, C.; Nuzillard, J.-M.; Verpoorte, R. J. Nat. Prod. 2006, 69, 742–748.
[https://doi.org/10.1021/np050535b]

-
Nagai, M.; Akita, K.; Yamada, K.; Okunishi, I. J. Nat. Med. 2010, 64, 305–312.
[https://doi.org/10.1007/s11418-010-0412-y]

-
Lu, C.-W.; Yeh, K.-C.; Chiu, K.-M.; Lee, M.-Y.; Lin, T.-Y.; Wang, S.-J. Int. J. Mol. Sci. 2022, 23, 8752.
[https://doi.org/10.3390/ijms23158752]

-
Dubois, M.-A.; Zoll, A.; Bouillant, M.-L.; Chopin, J. Phytochemistry 1982, 21, 1141–1143.
[https://doi.org/10.1016/S0031-9422(00)82433-4]

-
Narita, Y.; Inouye, K. Food Chem. 2011, 127, 1532–1539.
[https://doi.org/10.1016/j.foodchem.2011.02.013]

-
Dai, C.; Li, H.; Zhao, W.; Fu, Y.; Cheng, J. J. Anim. Physiol. Anim. Nutr. (Berl) 2024, 108, 439–450.
[https://doi.org/10.1111/jpn.13905]

-
Meyer, M. B.; Benkusky, N. A.; Sen, B.; Rubin, J.; Pike, J. W. J. Biol. Chem. 2016, 291, 17829–17847.
[https://doi.org/10.1074/jbc.M116.736538]

-
Yi, S. A.; Lee, J.; Park, S. K.; Kim, J. Y.; Park, J. W.; Lee, M. G.; Nam, K. H.; Park, J. H.; Oh, H.; Kim, S.; Han, J.; Kim, B. K.; Jo, D.-G.; Han, J.-W. J. Ginseng Res. 2020, 44, 58–66.
[https://doi.org/10.1016/j.jgr.2018.08.002]

-
Ciuffreda, M. C.; Malpasso, G.; Musarò, P.; Turco, V.; Gnecchi, M. In Mesenchymal Stem Cells: Protocols for in vitro Differentiation of Human Mesenchymal Stem Cells into Osteogenic, Chondrogenic and Adipogenic Lineages; Ciuffreda, M. C.; Malpasso, G.; Musarò, P.; Turco, V.; Gnecchi, M. Ed; Springer; New York, NY, USA, 2016, pp 149–158.
[https://doi.org/10.1007/978-1-4939-3584-0_8]

-
Kang, M.-H.; Lee, S.-J.; Lee, M.-H. J. Ginseng Res. 2020, 44, 823–832.
[https://doi.org/10.1016/j.jgr.2020.05.003]