Argutinic acid, A New Triterpenoid from the Fruits of Actinidia arguta
Abstract
Actinidia arguta, also called as kiwiberry or hardy kiwifruit, is one of the major varieties of kiwi plants. It is small in size and has no hairs on the surface, which making it easy to be consumed. In addition, it can be cultivated in Asia, due to its cold resistance. We have been interested in the beneficial effects of the fruits of A. arguta and investigated the constituents and biological activities. A new triterpenoid, argutinic acid, along with three triterpenoids and three megastigmines were isolated from the fruits of A. arguta. The structures were determined by spectroscopic analysis including NMR, MS, UV and IR. Among the isolated compounds, two triterpenes, argutinic acid (1) and usrolic acid (4) showed α-glucosidase inhibitory activities.
Keywords:
Actinidia arguta, triterpenoid, megastigmine, argutinic acid, α-glucosidaseIntroduction
The fruits of the genus Actinidia is well known as kiwi fruit and widely consumed as fresh fruits.1 Among diverse species of Actinidia, the fruit of A. arguta, which also called as kiwiberry or hardy kiwifruit, has distinct characteristics such as small and smooth-fruited and tolerance to low temperatures.2 It is rich in beneficial nutrients including organic acids, sugars, vitamins and minerals as well as bioactive compounds.3–4 In our previous research, we isolated diverse phenolic compounds, especially phenolic derivatives conjugated with different organic acids and evaluated the anti-inflammatory and antioxidant activities.5–6 In a continuation of our research on hardy kiwifruit, here we report the isolation and structural elucidation of seven compounds including a new triterpene and their anti-diabetic effect.
Experimental
Plant material – The fruits of A. arguta was collected at the National Institute of Forest Science in August 2016. After identification by the herbarium of the College of Pharmacy, Chungbuk National University, voucher specimens (CBNU2016-AA) were deposited in a specimen room of the herbarium.
General experimental – A Jasco UV-550 and Perkin-Elmer model LE599 spectrometer were used for the measurement of UV and IR spectra, respectively. NMR spectra were recorded on a Bruker DRX 400 or 500 MHz spectrometer using pyridine-d6 as a solvent. ESI-MS data were obtained on VG Autospec Ultima mass spectrometers using H2 as carrier gas with a Hypersil Gold column (flow rate 0.2 mL/min). HR-ESI-MS data was obtained using a maXis 4G (Bruker, Germany). Semipreparative HPLC was performed using a Waters HPLC A system equipped with Waters 600 Q-pumps, a 996 photodiode array detector, and Waters Empower software using a Gemini-NX ODS-column (5 μm, 10 × 150 mm).
Isolation of compounds – The dried powder of A. arguta fruits (12.0 kg) was extracted the EtOAc fraction (AAE, 31.3 g) was chromatographed on silica gel eluting with a mixture of CH2Cl2-MeOH by step gradient to give 12 subfractions (AAE1–AAE12). Subfraction AAE1 was subjected to Sephadex LH-20 eluting with n-hexane-CH2Cl2-MeOH (5:5:1) to give 10 subfractions (AAE1A–AAE1J). Compound 1 (2.6 mg) was obtained from AAE1A by MPLC on silica gel eluted with mixtures of n-hexane-EtOAc (5:1). Compounds 6 (4.6 mg) and 7 (3.9 mg) were purified from AAE1J by semi-preparative HPLC (Gemini-NX ODS-column, 5 μm, 10 × 150 mm) eluting with MeOH-water (20:80). Subfraction AAE110 was subjected to MPLC on RP-silica gel and eluted with mixtures of MeOH-H2O to obtain 7 subfractions (AAE10A-AAE10G). Further purification of AAE10A by Sephadex LH-20 eluting with CH2Cl2-MeOH (1:1) yielded compound 5 (6.1 mg). Subfraction AAE11 was subjected to MPLC on RP-silica gel and eluted with mixtures of MeOH-H2O to obtain 11 subfractions (AAE11A–AAE11K). Subfraction AAE11K was subjected to Sephadex LH-20 eluting with CH2Cl2-MeOH (1:1) and compounds 2 (2.6 mg), 3 (4.4 mg) and 4 (12.9 mg) were obtained by semi-preparative HPLC (Gemini-NX ODS-column, 5 μm, 10 × 150 mm) eluting with acetonitrile-water (23:77).
Argutinic acid (1) – brown syrup; −12.7 (c 0.1, MeOH); IR (KBr) νmax 3430, 1666 cm-1; ESIMS m/z 543 [M+Na]+; HRESIMS m/z 543.3292 ([M+Na]+ calcd. for C30H48NaO7 543.3298); 1H-NMR (500 MHz, pyridine-d5); δH 2.31 (1H, dd, J = 12.5, 4.0 Hz, H-1a), 1.39 (1H, m, H-1b), 4.25 (1H, m, H-2), 4.24 (1H, d, J = 10.5 Hz, H-3), 1.84 (1H, dd, J = 10.5, 1.5 Hz, H-5), 1.77 (1H, m, H-6), 1.38 (1H, m, H-7a), 1.75 (1H, m, H-7b), 1.91 (1H, dd, J = 10.5, 7.0 Hz, H-9), 1.72 (1H, m, H-11a), 2.07 (1H, m, H-11b), 2.06 (1H, m, H-15a), 2.85 (1H, ddd, J = 13.5, 10.0, 3.5 Hz, H-15b), 1.22 (1H, m, H-16a), 2.40 (1H, m, H-16b), 3.56 (1H, d, J = 12.0 Hz, H-18), 2.49 (1H, m, H-19), 2.48 (1H, m, H-21), 2.07 (1H, m, H-22a), 2.41 (1H, m, H-22b), 1.10 (3H, s, CH3-23), 1.09 (3H, s, CH3-25), 1.14 (3H, s, CH3-26), 1.28 (3H, s, CH3-27), 4.24 (1H, d, J = 10.5 Hz, H-24a), 3.76 (1H, d, J = 10.5 Hz, H-24b), 1.39 (3H, d, J = 7.5 Hz, CH3-29), 3.99 (1H, d, J = 10.5 Hz, H-30a), 4.18 (1H, d, J = 10.5 Hz, H-30b); 13C-NMR (125 MHz, pyridine-d5); δC 47.6 (C-1), 68.7 (C-2), 78.0 (C-3), 43.4 (C-4), 47.9 (C-5), 18.3 (C-6), 33.0 (C-7), 39.9 (C-8), 47.9 (C-9), 38.1 (C-10), 23.6 (C-11), 125.4 (C-12), 139.2 (C-13), 42.4 (C-14), 32.2 (C-15), 28.5 (C-16), 47.7 (C-17), 47.7 (C-18), 35.7 (C-19), 72.9 (C-20), 31.1 (C-21), 24.2 (C-22), 14.1 (C-23), 66.4 (C-24), 17.3 (C-25), 17.4 (C-26), 23.6 (C-27), 179.9 (C-28), 12.6 (C-29), 69.6 (C-30).
Measurement of α-glucosidase inhibitory activity – The inhibitory effect on α-glucosidase was measured using α-glucosidase from Saccharomyces cerevisiae (EC 3.2.1.20). A test sample was mixed with 80 μL enzyme buffer and 10 μL α-glucosidase and incubated for 15 min at 37oC. Then, after addition of 10 μL p-nitrophenyl α-D-glucopyranoside solution for enzyme reaction, the amount of p-nitrophenol cleaved by the enzyme was determined by measuring the absorbance at 405 nm in a 96-well microplate reader. Acarbose was used as a positive control.
Results and Discussion
Seven compounds were isolated from the fruits of A. arguta and the structures of known compounds were identified as three triterpenes, niga-ichigoside F2 (2), and niga-ichigoside F1 (3) and ursolic acid (4), and three magastigmanes, roseoside (5), vomifoliol (6) and loliolide (7), respectively, by comparing them with the literature (Fig. 1).6–9
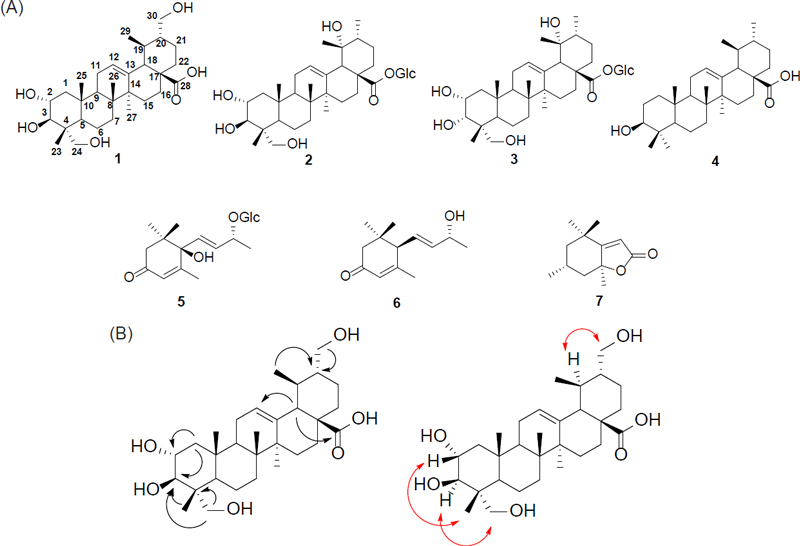
(A) Structures of compounds 1–7 from the fruits of A. arguta and (B) Key HMBC (→) and NOESY (↔) correlations of compound 1
Compound 1 was isolated as a brown amorphous powder, and the molecular formula was determined as C30H48O7 by HRESI-TOF-MS of molecular ion peak at m/z 543.3292 [M+Na]+ (calcd. for C30H48NaO7, 543.3298). Compound 1 was suggested as an ursane triterpenoid 6 from the one carboxyl group [δC 179.9 (C-28)], four tertiary methyl groups [δH 1.28 (3H, s, H-27), 1.14 (3H, s, H-26), 1.10 (3H, s, H-23), 1.09 (3H, s, H-25); δC 23.6 (C-27), 17.4 (C-26), 17.3 (C-25), 14.1 (C-23)], one secondary methyl group [δH 1.39 (3H, d, J = 7.0 Hz, H-29); δC 12.6 (C-29)], two oxymethine groups [δH 4.25 (1H, m, H-2), 4.24 (1H, d, J = 10.5 Hz, H-3); δC 68.7 (C-2), 78.0 (C-3)], one olefin group [δH 5.61 (1H, br s, H-12); δC 139.2 (C-13), 125.4 (C-12)] in 1H-NMR, 13C-NMR and HSQC spectra. Additionally, presence of two hydroxymethyl groups was suggested from the signals at [δH 4.24 (1H, d, J = 10.5 Hz, H-24a), 3.76 (1H, d, J = 10.5 Hz, H-24b); δC 66.4 (C-24)] and [δH 4.18 (1H, d, J = 10.5 Hz, H-30a), 3.99 (1H, d, J = 10.5 Hz, H-30b); δC 69.6 (C-30)]. These data are quite similar to those of compounds 2–4, suggesting that it is a ursolic acid derivatives. Compared to those of compound 2, additional presence of a hydroxymethyl group and absence of a glucose were deduced. The positions of these two hydroxymethyl groups were determined as C-24 and C-30 by the HMBC correlations of H24/C4 and H30/C20, respectively. The stereochemistry of 2-OH, 3-OH, 24-hydroxymethyl and 30- hydroxymethyl groups were determined as 2α, 3β, 24α and 30α by the J value (J2–3 = 10.5 Hz) and NOESY correlations between H2/H23, H3/H24 and H19/H30. Based on the above analysis results, the structure of compound 1 was determined as shown and named argutinic acid as it is first reported in nature.
As α-glucosidase is an important therapeutic in diabetes, 10–12 we measured the inhibitory effects of compounds from A. arguta fruits against α-glucosidase. Ursolic acid (4) exists in many plants and has already been reported to have α-glucosidase inhibitory activity. 13 Our results also showed an inhibitory effect of 60.2% inhibition at the concentration of 100 μM. The new triterpene, argutinic acid (1) also showed 44.2% inhibition against α-glucosidase at the concentration of 100μM. However, two triterpene glycosides (2 and 3) and three megastigmanes (5–7) exerted little effects.
Kiwi fruits, the fruits of the genus Actinidia, are widely consumed as fruits. It is rich in diverse bioactive ingredients especially phenolic compounds, and it is known to have excellent antioxidant effects. In this study, we conducted a study on the composition of the fruits of A. arguta and isolated one new triterpene, argutinic acid along with three triterpenes and three megastigmines. Triterpenes of A. arguta showed α-glucosidase inhibitory effects. Therefore, this study will contribute to the utilization of A. arguta fruits.
Acknowledgments
This work was supported by the Research grant (2022R1A2C1008081) of the National Research Foundation of Korea (NFR) funded by the Korean government (MSIT).
References
-
Latocha, P. Plant Foods Hum. Nutr. 2017, 72, 325–334.
[https://doi.org/10.1007/s11130-017-0637-y]

-
Sun, S.; Fang, J.; Lin, M.; Qi, X.; Chen, J.; Wang R.; Li Z.; Li Y.; Muhammad A. Plants 2020, 9, 515.
[https://doi.org/10.3390/plants9040515]

-
Pinto, D.; Delerue-Matos, C.; Rodrigues F. Food Res. Int. 2020, 136, 109449.
[https://doi.org/10.1016/j.foodres.2020.109449]

-
Zhang, H.; Teng, K.; Zang, H. Molecules 2023, 28, 7820.
[https://doi.org/10.3390/molecules28237820]

-
Ahn, J. H.; Park, Y.; Jo, Y. H.; Kim, S. B.; Yeon, S. W.; Kim, J. G.; Turk, A.; Song, J. Y.; Kim, Y.; Hwang, B. Y.; Lee, M. K. Food Chem. 2020, 308, 125666.
[https://doi.org/10.1016/j.foodchem.2019.125666]

-
Ahn, J. H.; Yeon, S. W.; Ryu, S. H.; Lee, S.; Turk, A.; Hwang, B. Y.; Lee, M. K. Phytochem. Lett. 2022, 48, 128–131
[https://doi.org/10.1016/j.phytol.2022.01.019]

-
Kim, Y. H.; Kang, S. S. Arch. Pharm. Res. 1993, 16, 109–113.
[https://doi.org/10.1007/BF03036856]

-
Gnoatto, S. C. B.; Dassonville-Klimpt, A.; Nascimento, S. D.; Galéra, P.; Boumediene, K.; Gosmann, G.; Sonnet, P.; Moslemi, S. Eur. J. Med. Chem. 2008, 43, 1865–1877.
[https://doi.org/10.1016/j.ejmech.2007.11.021]

-
Yajima, A.; Oono, Y.; Nakagawa, R.; Nukada, T.; Yabuta, G. Bioorg. Med. Chem. 2009, 17, 189–194.
[https://doi.org/10.1016/j.bmc.2008.11.002]

-
Ryu, S. H.; Kim, B. S.; Turk, A.; Yeon, S. W.; Lee, S.; Lee, H. H.; Ko, S. M.; Hwang, B. Y.; Lee, M. K. Nat. Prod. Sci. 2023, 29, 132–137.
[https://doi.org/10.20307/nps.2023.29.3.132]

-
Kuang, H.-X.; Yang, B.-Y.; Xia, Y.-G.; Feng, W.-S. Arch. Pharm. Res. 2008, 31, 1094–1097.
[https://doi.org/10.1007/s12272-001-1274-6]

-
Phong, N. V.; Thanh, L. T.; Kwon, M. J.; Lee, Y. M.; Min, B. S.; Kim, J. A. Nat. Prod. Sci. 2023, 29, 349–356.
[https://doi.org/10.20307/nps.2023.29.4.349]

-
Wang, J.; Zhao, J.; Yan, Y.; Liu, D.; Wang, C.; Wang, H. J. Sci. Food. Agric. 2020, 100, 986–994.
[https://doi.org/10.1002/jsfa.10098]