Cytotoxicity against MDA-MB-231 Breast Cancer Cells of Fungal Metabolites of Trichoderma sp. Collected from Medicinal Herbal Garden
Abstract
Trichoderma sp. isolated from the herbal garden is one of the well-known soil fungi, and various metabolites produced by this genus, such as anthraquinones, azaphilones and peptaibols, have been reported to exhibit cytotoxicity against various cancer cells. A large-scale cultivation and a chemical investigation of Trichoderma sp. led to isolation and purification of 10 known compounds from its EtOAc extract. Their structures were elucidated by comparing 1D NMR (1H and 13C) and HRESIMS data with previously reported literature. The cytotoxicity of all isolated compounds was measured against MDA-MB-231 breast cancer cells, and koninginin E (10) showed significant inhibitory activity with an IC50 value of 7.3 µM.
Keywords:
Soil fungus, Trichoderma sp., MDA-MB-231, Cytotoxicity, Koninginin EIntroduction
The plant sources of medicinal compounds are grown in the soil for years, where their roots play significant roles in various interactions with soil microorganisms to live and obtain nutrients.1,2 Indeed, metabolites of soil microorganisms themselves, have been widely reported to exhibit various bioactivities such as protecting plants from harmful pathogens, and have also been developed into clinical drugs, such as chloramphenicol (antibacterial), griseofulvin (antifungal), ivermectin and avermectin (antiparasitic), and cyclosporin A (immunosuppressant), which can be used to control various diseases in human.3–6
To find novel bioactive molecules, most natural product chemistry laboratories mainly collect bacterial or fungal strains from undisclosed and unique environments that cannot be easily approached such as the Antarctic or deep sea.7,8 So, our research group, Natural Products Drug Discovery laboratory (NPDD lab), is also collecting fungal strains from diverse soil environments across South Korea. The rationale behind this approach is that soil from different regions may experience various climatic conditions, such as precipitation and temperature, and support different plant species. Consequently, the metabolites of soil microorganisms interacting with these plants may also differ.9 Furthermore, some reports have shown that there are differences in microbial communities between ordinary soil and soil where medicinal plants are cultivated.10 For example, research has been conducted on the soil characteristics of regions famous for cultivating Panax ginseng, a representative medicinal plant of South Korea.11 Soils for growing high-quality ginseng were generally found at elevations between 200-700 meters, with pH levels ranging from 3.8 to 5.4, indicating slightly acidic to acidic conditions. The soil microbial community analysis revealed a relatively high abundance of Proteobacteria phylum. These findings suggest that the types of plants growing in the soil can influence the interacting microbial communities. Among our soil collection near Duksung Women’s University, we focused on soil from an herb garden where medicinal plants known for anti-cancer properties, such as Liriope platyphylla,12 Sedum kamtschaticum,13 Chrysanthemum zawadskii,14 and Chelidonium majus,15 grow annually. Through this approach, we aimed to discover cytotoxic metabolites from soil fungi symbiotic with these medicinal plants. Herein, we describe the isolation, purification, structural elucidation, and cytotoxic activity evaluation of fungal metabolites produced by Trichoderma sp. (DS2-7 strain) isolated from the herbal garden soil sample.
Experimental
General experimental procedures – Nuclear magnetic resonance (NMR) spectra were obtained on Varian NMR spectrometers (400 MHz for 1H and 100 MHz for 13C, Varian Inc., Palo Alto, CA, USA). The preparative HPLC was performed using Waters HPLC system equipped with pumps, a 996 photodiode-array detector (Waters Corporation, Milford, MA, USA), and a Luna 5 μm C18 column (100 Å, 250 × 21.2 mm I.D., Phenomenex, Torrance, CA, USA) with a flow rate of 10 mL/min. The semi-preparative HPLC were conducted on the same Waters HPLC system using a Luna 5 μm C18 (100 Å, 250 × 10 mm I.D., Phenomenex, Torrance, CA, USA) with a flow rate of 4 mL/min. The HPLC analysis was carried out on the same Waters HPLC system using a Luna 5 μm C18 (100 Å, 250 × 4.6 mm I.D., Phenomenex, Torrance, CA, USA) with a flow rate of 1 mL/min. HRESIMS analysis was performed using a UHPLC-HR-Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) system equipped with a diode array detector and an YMC-Triart C18 column (2.0 μm, 100 × 2.1 mm I.D. YMC Co., Ltd., Kyoto, Japan) with flow rate 0.3 mL/min. All solvents used were of ACS grade or better.
Fungal isolates and fermentation – Trichoderma sp. (Deposit No.: DS2-7) was isolated from a soil sample collected from the medicinal herbal garden in the College of Pharmacy of Duksung Women’s University, Seoul, South Korea (37.6510380, 127.0187136) in November 2022. Fungi were identified based on the ribosomal internal transcribed spacer (ITS) region (Macrogen, Korea). The resulting sequence data were compared to fungal sequences in GenBank, which revealed 100% identity matches to isolates described as Trichoderma sp. (GenBank accession no. KJ817310.1). The sequence data were deposited in GenBank (Trichoderma sp.: GenBank accession no. PQ340138).
To conduct large-scale cultivation for the isolation and purification of the cell cytotoxic metabolites, fungi were recovered from cryogenic storage (stored in a vial at −80oC as mycelium with 20% aqueous glycerol). Following recovery on rose bengal medium plates (15 g agar, 10 g malt extract, 1 g yeast, 0.05 g chloramphenicol, 0.025 g rose bengal, 1 L deionized H2O), fungal mycelia were aseptically cut into small pieces (~ 0.5 cm2) for the largescale fermentation inoculum. Large-scale fermentation was carried out using ten 1 L flasks with bilayers of Cheerios™ breakfast cereal as the medium, supplemented with a 0.3% sucrose solution and 0.005% chloramphenicol. Pieces of mycelia were aseptically inoculated into ten flasks and the cultures were grown at room temperature for three weeks.16,17
Extraction and isolation of Trichoderma sp. – The fungal culture was extracted with EtOAc (0.5 L × 3) at room temperature overnight. The extract was filtered and evaporated under reduced pressure using a rotary vacuum evaporator to obtain the EtOAc-soluble residue (DS2-7-A, 2.5 g). This residue was subjected to silica gel vacuum liquid column chromatography and eluted with dichloromethane 100% (DS2-7-B), dichloromethane-MeOH 9:1 (DS2-7-C), MeOH 100% (DS2-7-D). DS2-7-C (0.9 g) was separated using HP20ss gel vacuum liquid column chromatography and eluted with 30% MeOH in water (DS2-7-E), 50% MeOH (DS2-7-F), 70% MeOH (DS2-7-G), 90% MeOH (DS2-7-H), 100% MeOH (DS2-7-I), dichloromethane-MeOH 1:1 (DS2-7-J). DS2-7-G (58 mg) was subjected to preparative HPLC (C18, gradient from 60% to 75% MeOH in H2O over 20 min, flow rate of 10 mL/min) to obtain 8 subfractions (DS2-7-G-1~8). Among these subfractions, DS2-7-G-7 and DS2-7-G-8 were turned out to be compounds 9 (4.5 mg) and 10 (3.2 mg), respectively. DS2-7-G-1 (6.3 mg) was subjected to semi-preparative HPLC (C18, isocratic 25% aqueous MeCN, flow rate: 4 mL/min) to give compound 8 (3.5 mg, tR = 16.4 min). DS2-7-G-2 (3.0 mg) was purified by semi-preparative HPLC (C18, isocratic 20% aqueous MeCN, flow rate: 4 mL/min) to give compound 1 (0.9 mg, tR = 26.0 min). DS2-7-G-4 (5.8 mg) was subjected to semi-preparative HPLC (C18, isocratic 22% aqueous MeCN, flow rate: 4 mL/min), yielding compounds 2 (0.9 mg, tR = 12.9 min), 3 (0.2 mg, tR = 13.6 min) and 4 (2.0 mg, tR = 22.2 min). Compound 5 (3.7 mg, tR = 16 min) was isolated from DS2-7-H (33 mg) using semi-preparative HPLC (C18, isocratic 50% aqueous MeCN, flow rate: 4 mL/min). DS2-7-I (55 mg) was separated by semi-preparative HPLC (C18, isocratic 25% aqueous MeCN, flow rate: 4 mL/min) to obtain 4 subfractions (DS2-7-I-1~4). DS2-7-I-2 (6.4 mg) was purified by semi-preparative HPLC (C18, isocratic 83% aqueous MeCN, flow rate: 4 mL/min) to obtain compounds 7 (1.0 mg, tR = 19.3 min) and 6 (1.0 mg, tR = 22.6 min).
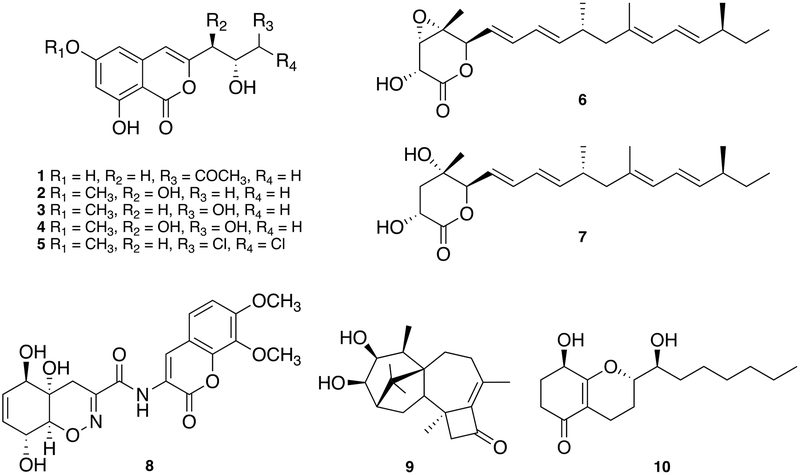
(-)-Citreoisocoumarin (1) – white powder; [α]D25−16 (c 0.1, MeOH); UV γmax (log ε): 236 sh, 244 (2.59), 256 sh, 275 sh, 325 (0.31) nm; HR-ESI-MS: m/z 301.0682 [M+Na]+ (calcd. for C14H14O6Na, 301.0683); 1H-NMR (CD3OD, 400 MHz): δ 6.33 (1H, s, H-5), 6.24 (1H, br s, H-4), 6.22 (1H, br s, H-7), 4.41 (1H, m, H-2ʹ), 2.67 (2H, m, H-3ʹ), 2.61 (2H, m, H-1ʹ), 2.16 (3H, s, H-5ʹ).
(+)-Peyroisocoumarin D (2) – white powder; [α]D25+30 (c 0.1, MeOH); UV γmax (log ε): 238 sh, 244 (3.18), 256 sh, 277 (0.42), 286 sh, 326 (0.37) nm; HR-ESI-MS: m/z 267.0862 [M+H]+ (calcd. for C13H15O6, 267.0863); 1H-NMR (CD3OD, 400 MHz): δ 6.61 (1H, br s, H-5), 6.51 (1H, br s, H-4), 6.46 (1H, br s, H-7), 4.14 (1H, m, H-9), 3.99 (1H, m, H-10) 3.86 (3H, s, 6-OCH3), 1.23 (3H, d, J = 6.4 Hz, H-11).
(+)-Diaportinol (3) – yellowish solid; [α]D25+10 (c 0.1, MeOH); UV γmax (log ε): 238 sh, 244 (1.61), 255 sh, 276 sh, 288 sh, 372 (0.19) nm; HR-ESI-MS: m/z 267.0862 [M+H]+ (calcd. for C13H15O6, 267.0863); 1H-NMR (CD3OD, 400 MHz): δ 6.47 (1H, s, H-5), 6.45 (1H, s, H-4), 6.45 (1H, s, H-7), 3.99 (1H, m, H-10), 3.85 (3H, s, OCH3), 3.54 (1H, d, J = 5.4 Hz, H-11), 2.76 (1H, dd, J = 14.8, 4.0, H-9), 2.53 (1H, dd, J = 14.8, 8.8 Hz, H-9).
(+)-9-Hydroxydiaportinol (4) – white solid powder; [α]D25 +22 (c 0.1, MeOH); UV γmax (log ε): 237 sh, 244 (2.03), 256 sh, 277 sh, 287 sh, 328 (0.25) nm; HR-ESIMS: m/z 283.0816 [M+H]+ (calcd. for C13H15O7, 283.0812); 1H-NMR (CD3OD, 400 MHz): δ 6.62 (1H, s, H-4), 6.54 (1H, s, H-5), 6.49 (1H, s, H-7), 4.32 (1H, d, J = 8.0 Hz, H-9), 4.06 (1H, m, H-10), 3.86 (3H, s, OCH3), 3.84 (1H, dd, J = 11.6, 2.8 Hz, H-11), 3.76 (1H, dd, J = 11.6, 6.0 Hz, H-11).
(+)-Dichlorodiaportin (5) – brownish solid; [α]D25 +14 (c 0.1, MeOH); UV γmax (log ε): 239 sh, 244 (1.42), 255 sh, 277 (1.81), 288 sh, 325 (1.74) nm; HR-ESI-MS: m/z 319.0135 [M+H]+ (calcd. for C13H13Cl2O5, 319.0135); 1H-NMR (CD3OD, 400 MHz): δ 6.51(2H, br s, H-4,5), 6.47 (1H, br s, H-7), 6.03 (1H, br s, H-11), 4.30 (1H, m, H-10), 3.86 (3H, s, 6-OCH3), 2.98 (1H, br d, J = 14.8 Hz, H-9), 2.74 (1H, dd, J = 14.8, 9.2 Hz, H-9); 13C-NMR (CD3OD, 100 MHz): δ 167.2 (C-6), 166.1 (C-1), 163.3 (C-8), 153.5 (C-3), 139.5 (C-4a), 106.4 (C-4), 100.9 (C-5) , 100.2 (C-7), 99.4 (C-8a), 75.8 (C-11), 72.8 (C-10), 54.9 (O-Me), 35.9 (C-9).
(+)-Nafuredin (6) – white powder; +32 (c 0.1, MeOH); UV γmax (log ε): 231 sh, 240 (0.86) nm; HR-ESIMS: m/z 361.2374 [M+H]+ (calcd. for C22H33O4, 361.2373); 1H-NMR (CD3OD, 400 MHz): δ 6.42 (1H, dd, J = 15.2, 10.2 Hz, H-7), 6.16 (1H, dd , J = 15.2, 10.2 Hz, H-14), 6.09 (1H, dd, J = 15.2, 10.2 Hz, H-8), 5.77 (1H, dd, J = 15.2, 7.6 Hz, H-9), 5.73 (1H, d, J = 10.0 Hz, H-13), 5.67 (1H, dd, J = 15.2, 8.4 Hz, H-6), 5.38 (1H, dd, J = 15.2, 7.6 Hz, H-15), 4.91 (1H, d, J = 8.4 Hz, H-5), 4.66 (1H, s, H-2), 3.45 (1H, s, H-3), 2.43 (1H, m, H-10), 2.06 (1H, m, H-11), 2.03 (1H, m, H-16), 1.99 (1H, m, H-11), 1.69 (3H, s, H-21), 1.39 (3H, s, H-19), 1.29 (2H, m, H-17), 0.97 (3H, d, J = 6.8 Hz, H-22), 0.96 (3H, d, J = 6.8 Hz, H-20), 0.85 (3H, t, J = 7.2 Hz, H-18); 13C-NMR (CD3OD, 100 MHz): δ 170.7 (C-1), 143.7 (C-9), 137.9 (C-15), 137.5 (C-7), 133.4 (C-12), 126.7 (C-13), 126.6 (C-8), 124.7 (C-14), 123.2 (C-6), 80.1 (C-5), 67.5 (C-2), 59.4 (C-3), 58.0 (C-4), 47.0 (C-11), 38.6 (C-16), 34.8 (C-10), 29.5 (C-17), 19.3 (C-22), 18.6 (C-20), 16.4 (C-19) 15.1 (C-21), 10.7 (C-18).
(+)-Nafuredin C (7) – yellowish gum; [α]D25 +12 (c 0.1, MeOH); UV γmax (log ε): 231 sh, 240 (2.30) nm; HR-ESIMS: m/z 385.2352 [M+Na]+ (calcd. for C22H34O4Na, 385.2349); 1H-NMR (CD3OD, 400 MHz): δ 6.30 (1H, dd, J = 15.2, 10.4 Hz, H-7), 6.16 (1H, dd, J = 15.2, 10.4 Hz, H-14), 6.03 (1H, dd, J = 15.2, 10.4 Hz, H-8), 5.73 (1H, d, J = 10.8 Hz, H-13), 5.63 (1H, dd, J = 15.2, 7.6 Hz, H-9), 5.52 (1H, dd, J = 15.2, 7.6 Hz, H-6), 5.37 (1H, dd, J = 15.2, 7.6 Hz, H-15), 4.56 (1H, m, H-2), 4.10 (1H, d, J = 6.8 Hz, H-5), 2.40 (1H, m, H-10), 2.23 (2H, br d, J = 9.2 Hz, H-3), 2.05 (1H, m, H-11), 2.03 (1H, m, H-16), 1.98 (1H, m, H-11), 1.68 (3H, s, H-21), 1.30 (3H, s, H-19), 1.29 (2H, m, H-17), 0.97 (3H, t, J = 7.2 Hz, H-22), 0.95 (3H, t, J = 7.2 Hz, H-20), 0.84 (3H, t, J = 7.2 Hz, H-18); 13C-NMR (CD3OD, 100 MHz): δ 177.3 (C-1), 141.1 (C-9), 137.8 (C-15), 134.0 (C-7), 133.6 (C-12), 127.5 (C-8), 127.3 (C-6), 126.6 (C-13), 124.8 (C-14), 85.0 (C-4), 75.9 (C-5), 67.6 (C-2), 47.2 (C-11), 38.7 (C-16), 36.5 (C-3), 34.8 (C-10), 29.5 (C-17), 21.6 (C-19), 19.4 (C-22), 18.9 (C-20), 15.1 (C-21), 10.7 (C-18).
(+)-Trichodermamide A (8) – colorless solid; [α]D25 +80 (c 0.1, MeOH); UV γmax (log ε): 242 (0.58), 345 (1.68) nm; HRESIMS: m/z 433.1243 [M+H]+ (calcd. for C20H21N2O9, 433.1242); 1H-NMR (CD3OD, 400 MHz): δ 8.60 (1H, s, H-3ʹ), 7.31 (1H, d, J = 8.8 Hz, H-5ʹ), 7.15 (1H, d, J = 8.8Hz, H-6ʹ), 5.60 (1H, d, J = 10.8 Hz, H-6), 5.52 (1H, d, J = 10.8 Hz, H-7), 4.37 (1H, m, H-5), 4.12 (1H, m, H-8), 4.10 (1H, m, H-9), 3.93 (3H, s, 7ʹ-OCH3), 3.91 (3H, s, 8ʹ- OCH3), 2.67 (1H, dd, J = 19.6, 1.6 Hz, H-3), 2.24 (1H, d, J = 19.6 Hz, H-3); 13C-NMR (CD3OD, 100 MHz): δ 161.3 (C-1), 158.2 (C-1ʹ), 154.2 (C-7ʹ), 150.0 (C-2), 143.9 (C-9ʹ), 135.8 (C-8ʹ), 129.7 (C-6), 127.3 (C-7), 123.9 (C-3ʹ), 122.5 (C-5ʹ), 121.0 (C-2ʹ), 113.9 (C-4ʹ), 109.6 (C-6'), 83.6 (C-9), 73.6 (C-5), 67.8 (C-4), 66.8 (C-8), 60.3 (8ʹ-OCH3), 55.5 (7ʹ-OCH3), 22.7 (C-3).
(+)-Harzianol E (9) – white powder; [α]D25 +16 (c 0.1, MeOH); UV γmax (log ε): 260 (2.36) nm; HR-ESI-MS: m/z 341.2074 [M+Na]+ (calcd. for C20H30O3Na, 341.2087); 1H-NMR (CD3OD, 400 MHz): δ 3.61 (1H, m, H-3), 3.60 (1H, m, H-4), 2.54 (1H, d, J = 16.2 Hz, H-12), 2.45 (1H, m, H-8), 2.39 (1H, m, H-5), 2.32 (1H, d, J = 16.2 Hz, H-12), 2.04 (3H, s, H-20), 2.02 (1H, m, H-8), 2.00 (2H, m, H-14,15), 1.92 (1H, dd, J = 14.0, 7.2 Hz, H-7), 1.68 (1H, m, H-2), 1.46 (3H, s, H-19), 1.21 (3H, d, J = 7.2 Hz, H-18), 1.20 (1H, m, H-7), 1.16 (1H, m, H-15), 1.00 (3H, s, H-17), 0.86 (3H, s, H-16); 13C-NMR (CD3OD, 100 MHz): δ 199.1 (C-11), 150.2 (C-10), 147.6 (C-9), 83.3 (C-3), 81.6 (C-4), 58.8 (C-12), 51.1 (C-14), 50.5 (C-6), 49.3 (C-2), 45.5 (C-1), 39.0 (C-13), 37.7 (C-5), 30.0 (C-8), 28.0 (C-7), 27.2 (C-15), 24.6 (C-16), 22.3 (C-17), 20.4 (C-20), 19.8 (C-18), 19.6 (C-19).
(+)-Koninginin E (10) – white powder; [α]D25 +8 (c 0.1, MeOH); UV γmax (log ε): 262 (4.20) nm; HR-ESI-MS: m/z 283.1903 [M+H]+ (calcd. for C16H27O4, 283.1904); 1H-NMR (CD3OD, 400 MHz): δ 4.33 (1H, d, J = 4.8 Hz, H-4), 3.85 (1H, ddd, J = 10.4, 4.4, 2.0 Hz, H-9), 3.63 (1H, m, H-10), 2.60 (1H, ddd, J = 16.8, 9.6, 4.8 Hz, H-2), 2.37 (1H, d, J = 16.8, 5.2 Hz, H-7), 2.26 (1H, m, H-2), 2.13 (1H, m, H-3), 2.06 (1H, m, H-7), 1.93 (2H, m, H-3 and H-8), 1.62 (1H, m, H-8), 1.58 (2H, m, H-11), 1.25–1.55 (8H, m, C-12,13,14,15), 0.88 (3H, t, J = 6.8 Hz, H-16); 13C-NMR (CD3OD, 100 MHz): δ 198.9 (C-1), 171.2 (C-5), 110.8 (C-6), 80.8 (C-9), 72.2 (C-10), 65.1 (C-4), 32.1 (C-11), 32.0 (C-2), 31.5 (C-14), 29.0 (C-13), 28.9 (C-3), 25.2 (C-12), 22.2 (C-8), 22.1, (C-15), 17.3 (C-7), 12.9 (C-16).
Cell Viability Assay – Cell viability was determined using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Cells were seeded in 96-well plates and incubated at 37oC for 24 h. Subsequently, cells were treated with various concentrations of the isolated compounds (0, 1, 5, 10, 20, and 40 μM) for 24 h. After treatment, 25 μL of 5 mg/mL MTT (Sigma-Aldrich, St. Louis, MO, USA) was added to each well and incubated for 3 h. The formazan crystals were dissolved using 100 μL of dimethyl sulfoxide. The optical density was measured at 540 nm using a Synergy 2 Multi-Mode Reader (BioTek Instruments Inc., Winooski, VT, USA).18
Results and Discussion
An in vitro cytotoxicity screening was performed using MDA-MB-231 (breast cancer cells) on the 96 fungal extracts collected from the territory of inside or near Duksung Women’s University. A subset of 20 fungal strains was isolated from the herbal garden in the College of Pharmacy of Duksung Women’s University. Among the strains isolated from the herbal garden (DS2), the EtOAc extract of DS2-7 (46% inhibition at 10 μg/mL vs DMSO) exhibited relatively better activity against MDA-MB-231 cancer cells compared to the other strains (Fig. 1). Additionally, the HPLC analysis of the extract using our in-house method revealed several compounds that had not been dereplicated (Fig. 2). Therefore, we selected DS2-7, which was identified as Trichoderma sp., for large-scale fermentation to find cytotoxic compounds.
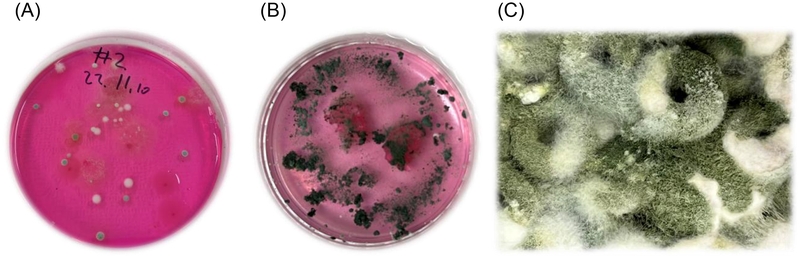
Pictures of fungi. (A) Soil fungi on RBM medium (DS2) isolated from herbal garden. (B) Trichoderma sp. DS2-7 on RBM medium isolated from DS2. (C) Trichoderma sp. DS2-7 cultivated on Cheerios breakfast cereal.
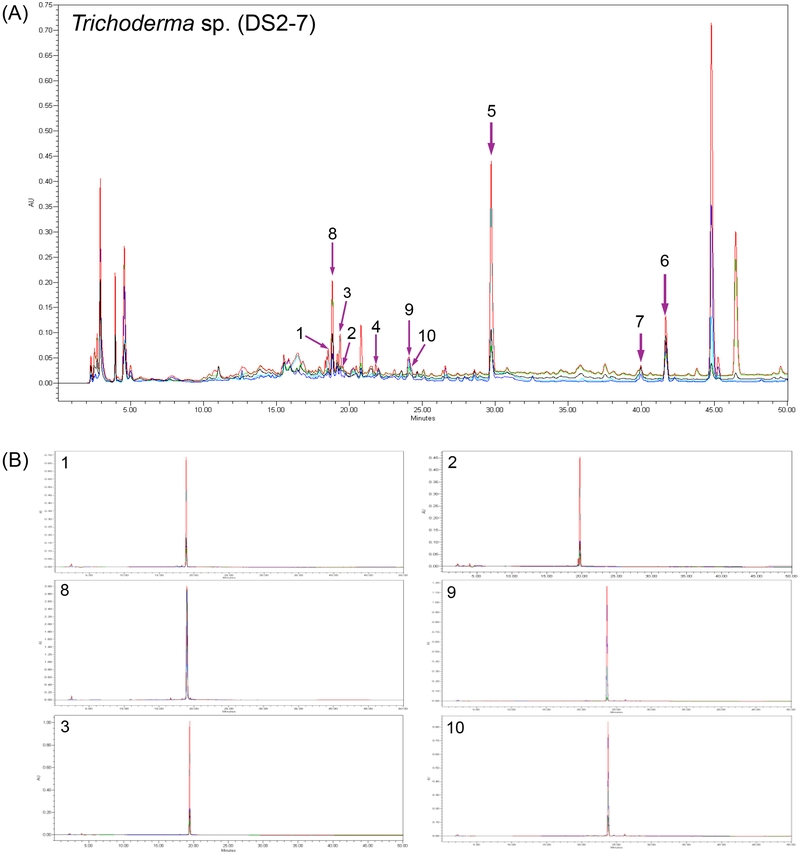
(A) HPLC chromatogram of EtOAc extract of Trichoderma sp. (DS2-7). (B) HPLC chromatograms of compounds 1–3 and 8–10.
Using a couple of vacuum liquid chromatography with silica gel and HP20ss gel, a total of 10 compounds were isolated from the large-scale fermentation of Trichoderma sp. DS2-7. Their absolute structures of the isolated compounds (1–10) were determined by comparing their spectroscopic data (1Hand 13C NMR, ROESY spectrum, optical rotation and HRESIMS data) with previously reported data (Fig. 3). They were identified as (-)-citreoisocoumarin (1)19 (+)- peyroisocoumarin (2),20 (+)-diaportinol (3),21 (+)-9- hydroxydiaportinol (4),22 (+)-dichlorodiaportin (5),21 (+)- nafuredin (6),23 (+)-nafuredin C (7),24 (+)-trichodermamide A (8),25 (+)-harzianol E (9),26 and (+)-koninginin E (10),27 respectively. These compounds included five isocoumarins (1–5), two nafuredin polyketides (6 and 7), a modified dipeptide (8), a diterpene (9) and a koninginin series polyketide (10), all of which were known metabolites produced by the genus Trichoderma.28
All isolated compounds were evaluated for their inhibitory activity against MDA-MB-231 (triple-negative breast cancer) cells with doxorubicin utilized as a positive control (IC50: 86.53 μM). Among them, compound 10 exhibited strong inhibitory activity with an IC50 of 7.3 μM (Table 1). Compounds 1, 6, and 7 exhibited inhibitory activities with IC50 values of 93.85, 87.10 and 109.64 μM, respectively, similar to doxorubicin (IC50: 86.53 μM), a clinically used anticancer agent, which was used as a positive control.
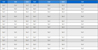
Cell viability assay results against MDA-MB-231 cancer cells (IC50 values expressed in μM ± SEM)a of compounds 1–10
In this study, we report in vitro cell viability screening, isolation of soil fungus, purification and structure elucidation of 10 fungal metabolites (1–10) from Trichoderma sp. and their cytotoxic activity against MDA-MB-231 breast cancer cells. Trichoderma sp. is one of the abundant fungi in soil that play numerous roles in promoting plant growth. These species inhibit the growth of pathogens, enhance plant immunity against oxidative stress, and improve soil conditions by facilitating the decomposition of organic matter.29–31 Trichoderma sp. has been known to produce a diverse range of secondary metabolites, including various polyketides, terpenoids, and peptaibols, which have been reported to exhibit various biological activities.32–34 Among these, koninginins, a group of polyketides, are notable for their anti-inflammatory effects and plant growth-regulating abilities.27,35 Our research has uncovered a remarkable cytotoxicity of koninginin E (10) against MDA-MB-231 cells. Moving forward, we plan to conduct further investigation focused on discovering new koninginin derivatives from soil fungi and exploring other bioactivities.
Acknowledgments
This research was supported by Duksung Women’s University Research Grants 2023-3000008765.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
REFERENCES
-
Jacoby, R.; Peukert, M.; Succurro, A.; Koprivova, A.; Kopriva, S. Front. Plant Sci. 2017, 8, 1617.
[https://doi.org/10.3389/fpls.2017.01617]

-
Tyc, O.; Song, C.; Dickschat, J. S.; Vos, M.; Garbeva, P. Trends Microbiol. 2017, 25, 280–292.
[https://doi.org/10.1016/j.tim.2016.12.002]

-
Feder Jr, H. M.; Osier, C.; Maderazo, E. G. Rev. Infect. Dis. 1981, 3, 479–491.
[https://doi.org/10.1093/clinids/3.3.479]

-
Petersen, A. B.; Rønnest, M. H.; Larsen, T. O.; Clausen, M. H. Chem. Rev. 2014, 114, 12088–12107.
[https://doi.org/10.1021/cr400368e]

-
Omura, S.; Crump, A. Nat. Rev. Microbiol. 2004, 2, 984–989.
[https://doi.org/10.1038/nrmicro1048]

-
Borel, J. F.; Feurer, C.; Gubler, H. U.; Stähelin, H. Agents Actions 1976, 6, 468–475.
[https://doi.org/10.1007/BF01973261]

-
Bracegirdle, J.; Casandra, D.; Rocca, J. R.; Adams, J. H.; Baker, B. J. J. Nat. Prod. 2022, 85, 2454–2460.
[https://doi.org/10.1021/acs.jnatprod.2c00684]

-
Cheng, M.; Tang, X.; Shao, Z.; Li, G.; Yao, Q. J. Nat. Prod. 2023, 86, 2342–2347.
[https://doi.org/10.1021/acs.jnatprod.3c00534]

-
Köberl, M.; Schmidt, R.; Ramadan, E. M.; Bauer, R.; Berg, G. Front. Microbiol. 2013, 4, 400.
[https://doi.org/10.3389/fmicb.2013.00400]

-
Li, L.; Yang, X.; Tong, B.; Wang, D.; Tian, X.; Liu, J.; Chen, J.; Xiao, X.; Wang, S. Front. Microbiol. 2023, 14, 1078886.
[https://doi.org/10.3389/fmicb.2023.1078886]

-
Kim, K.; Um, Y.; Jeong, D. H.; Kim, H.-J.; Kim, M. J.; Jeon, K. S. Korean J. Environ. Biol. 2019, 37, 380–388.
[https://doi.org/10.11626/KJEB.2019.37.3.380]

-
Wang, H.-C.; Wu, C.-C.; Cheng, T.-S.; Kuo, C.-Y.; Tsai, Y.-C.; Chiang, S.-Y.; Wong, T.-S.; Wu, Y.-C.; Chang, F.-R. Evid. Based Complement. Alternat. Med. 2013, 2013, 857929.
[https://doi.org/10.1155/2013/857929]

-
Aiello, P.; Sharghi, M.; Mansourkhani, S. M.; Ardekan, A. P.; Jouybari, L.; Daraei, N.; Peiro, K.; Mohamadian, S.; Rezaei, M.; Heidari, M.; Peluso, I.; Ghorat, F.; Bishayee, A.; Kooti, W. Oxid. Med. Cell. Longev. 2019, 2019, 2075614.
[https://doi.org/10.1155/2019/2075614]

-
Kim, K.-Y.; Oh, T.-W.; Yang, H.-Y.; Kim, Y.-W. Ma, J.-Y.; Park, K.-I. BMC Complement. Altern. Med. 2019, 19, 274.
[https://doi.org/10.1186/s12906-019-2688-0]

-
Maji. A. K.; Banerji, P. Int. J. Herb. Med. 2015, 3, 10–27.
[https://doi.org/10.22271/flora.2015.v3.i1.03]

-
Lee, J. W.; Collins, J. E.; Hulverson, M. A.; Aguila, L. K. T.; Kim, C. M.; Wendt. K. L.; Chakrabarti, D.; Ojo, K. K.; Wood, G. E.; Voorhis, W. C. V.; Cichewicz, R. H. J. Nat. Prod. 2023, 86, 1596–1605.
[https://doi.org/10.1021/acs.jnatprod.3c00283]

-
Lee, J. W.; Collins, J. E.; Wendt, K. L.; Chakrabarti, D.; Cichewicz, R. H. J. Nat. Prod. 2021, 84, 503–517.
[https://doi.org/10.1021/acs.jnatprod.0c01370]

-
Kim, E.-S.; Moon, A. Oncol. Lett. 2015, 9, 897–902.
[https://doi.org/10.3892/ol.2014.2735]

-
Lai, S.; Shizuri, Y.; Yamamura, S.; Kawai, K.; Furukawa, H. Heterocycles 1991, 32, 297–305.
[https://doi.org/10.3987/COM-90-5639]

-
Zhao, Y.; Liu, D.; Proksch, P.; Yu, S.; Lin, W. Chem. Biodivers. 2016, 13, 1186–1193.
[https://doi.org/10.1002/cbdv.201600012]

-
Larsen, T. O.; Breinholt, J. J. Nat. Prod. 1999, 62, 1182–1184.
[https://doi.org/10.1021/np990066b]

-
Ding, Z.; Tao, T.; Wang, L.; Zhao, Y.; Huang, H.; Zhang, D.; Liu, M.; Wang, Z.; Han, J. J. Microbiol. Biotechnol. 2019, 29, 731–738.
[https://doi.org/10.4014/jmb.1901.01015]

-
Ui, H.; Shiomi, K.; Yamaguchi, Y.; Masuma, R.; Nagamitsu, T.; Takano, D.; Sunazuka, T.; Namikoshi, M.; Omura, S. J. Antibiot. 2001, 54, 234–238.
[https://doi.org/10.7164/antibiotics.54.234]

-
Zhao, D.-L.; Zhang, X.-F.; Huang, R.-H.; Wang, D.; Wang, X.-Q.; Li, Y.-Q.; Zheng, C.-J.; Zhang, P.; Zhang, C.-S. Front. Microbiol. 2020, 11, 1495.
[https://doi.org/10.3389/fmicb.2020.01495]

-
Garo, E.; Starks, C. M.; Jensen, P. R.; Fenical, W.; Lobkovsky, E.; Clardy, J. J. Nat. Prods. 2003, 66, 423–426.
[https://doi.org/10.1021/np0204390]

-
Zhang, M.; Liu, J.; Chen, R.; Zhao, J.; Xie, K.; Chen, D.; Feng, K.; Dai, J. Tetrahedron 2017, 73, 7195–7199.
[https://doi.org/10.1016/j.tet.2017.11.002]

-
Parker, S. R.; Cutler, H. G.; Schreiner, P. R. Biosci. Biotech. Biochem. 1995, 59, 1126–1127.
[https://doi.org/10.1271/bbb.59.1126]

-
Li, W.; Xu, J.; Li, F.; Xu, L.; Li, C. Pharmacogn. Mag. 2016, 12, 259–261.
[https://doi.org/10.1163/156853211X00134]

-
Yao, X.; Guo, H.; Zhang, K.; Zhao, M.; Ruan, J.; Chen, J. Front. Microbiol. 2023, 14, 1160551.
[https://doi.org/10.3389/fmicb.2023.1160551]

-
Tyśkiewicz, R.; Nowak, A.; Ozimek, E.; Jaroszuk- Ściseł, J. Int. J. Mol. Sci. 2022, 23, 2329.
[https://doi.org/10.3390/ijms23042329]

-
Hermosa, R.; Viterbo, A.; Chet, I.; Monte, E. Microbiology 2012, 158, 17–25.
[https://doi.org/10.1099/mic.0.052274-0]

-
Park, Y. S.; Kim, E.-S.; Deyrup, S. T.; Lee, J. W.; Shim, S. H. J. Nat. Prod. 2024, 87, 2081–2094.
[https://doi.org/10.1021/acs.jnatprod.4c00590]

-
Han, J. S.; Kim, E.-S.; Cho, Y. B.; Kim, S. Y.; Lee, M. K.; Hwang, B. Y.; Lee, J. W. J. Nat. Prod. 2024, 87, 1994–2003.
[https://doi.org/10.1021/acs.jnatprod.4c00438]

-
Reino, J. L.; Guerrero, R. F.; Hernández-Galán, R.; Collado, I. G. Phytochem. Rev. 2008, 7, 89–123.
[https://doi.org/10.1007/s11101-006-9032-2]

-
Souza, A. D. L.; Rodrigues-Filho, E.; Souza, A. Q. L.; Pereira, J. O.; Calgarotto, A. K.; Maso, V.; Marangoni, S.; Silva, S. L. D. Toxicon. 2008, 51, 240–250.
[https://doi.org/10.1016/j.toxicon.2007.09.009]