Prunus yedoensis Bark Water Extract Increases Hyaluronic Acid Synthesis and Skin Barrier-related Protein Levels
Abstract
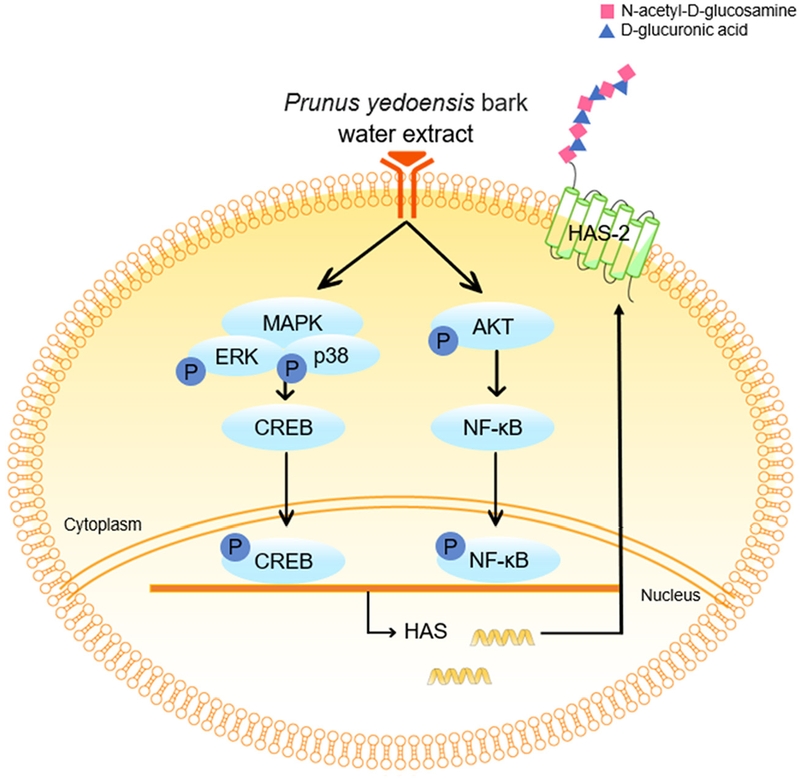
Damage to barrier function due to a loss of moisture in the skin can lead to skin diseases, such as atopy and psoriasis. Hyaluronic acid (HA) has the ability to retain large amounts of moisture and thereby protects the skin against moisture loss. In addition, tight junctions prevent water loss and maintain barrier functions by restricting molecular movement through cell-to-cell connections. In this study, the effects of Prunus yedoensis bark water extract (PYBWE) on HA synthesis and tight junction protein expression in HaCaT cells were evaluated. In HaCaT cells, PYBWE had effects on the MAPKs, CREB, AKT, NF-κB, and CLDN-4 signaling pathways, at 25, 50, and 100 μg/mL concentrations increasing the amount of HA produced via HAS-2 and effectively improving skin barrier function.
Keywords:
Prunus yedoensis Matsumura, Tight junction, Hyaluronic acid, MAPKs, HaCaT cellsIntroduction
The skin acts as a protective barrier surrounding the body and is composed of an epideral layer, dermal layer, and subcutaneous tissue.1 The epidermal layer is mainly composed of keratinocytes and contributes to protection against external factors, such as infectious pathogens, ultraviolet rays, and chemicals.2–4 This protective effect is mediated by two main physical barriers, the stratum corneum and tight junctions.5
Hyaluronic acid (HA) is a natural moisturizing factor with important roles not only in skin elasticity and the maintenance of moisture but also in intercellular interactions and signaling.6 HA is a high-molecular-weight linear glycosaminoglycan composed of repeating units of N-acetyl-D-glucosamine and D-glucuronic acid and is synthesized inside the cell membrane via three enzymes, hyaluronic acid synthase-1 (HAS-1), hyaluronic acid synthase-2 (HAS-2), and hyaluronic acid synthase-3 (HAS-3).7 Among HASs, HAS-1 has the lowest activity; however, HAS-2 and HAS-3 play important roles in various physiological and pathological processes.8 Water loss in the skin leads to dryness, which enables the penetration of external substances and can lead to allergic diseases as well as atopic dermatitis.9 Recent findings suggest that HAS gene expression controls HA synthesis, improves skin moisture, and reinforces skin barrier function.10
Tight junctions are cellular connection structures that can be classified into gap junction, desmosome junction, and adhesive junction; they are found mainly between epithelial and endothelial cells.11 Tight junctions in the skin seal the gap between epithelial cells in the stratum granulosum layer, which not only restricts material movement from the outside but also prevents the evaporation of moisture, thereby playing an important role in maintaining moisture in the skin.12 Tight junctions consist of three types of transmembrane proteins, occludin, claudins, and junctional adhesion molecules (JAMs), and are linked to actin in the cytoskeleton by the zonula occludins (ZO) protein to strengthen the structure.13 In particular, claudin plays the most important role among the tight junction proteins. It is composed of four transmembrane proteins arranged side-by-side in a linear manner to form tightly moving strands through lateral binding.14 In skin epithelial cells, CLDN-1 and CLDN-4 play important roles in skin barrier function and in maintaining the water barrier.15 In particular, CLDN-4 is frequently expressed in the stratum corneum, and a loss of tight junctions due to a reduction in CLDN-1 can lead to an imbalance in skin barrier function, leading to skin inflammatory diseases, such as atopic dermatitis; therefore, strengthening the skin barrier is important.16,17
A recent study has shown that the treatment of skin keratinocytes with HA improves skin barrier function by increasing the levels of tight junction proteins.18 The expression HAS-2, which is responsible for HA synthesis in the skin, depends on several signaling pathways, such as mitogen-activated protein kinase (MAPK), CREB, AKT, and NF-κB.19–21 Therefore, the expression of HAS-2 is regulated by interactions among various signaling pathways. These findings suggest that skin HA may play a key role in skin barrier function.
Prunus yedoensis(king cherry) is a tree species that has been used for traditional pharmaceutical and medical purposes for many years; the chemical and medical characteristics of its bark have been studied.22 The species has immunosuppressive and anti-inflammatory effects.23 Thus, P. yedoensis has a variety of physiologic effects on the skin.
We confirmed that the Prunus yedoensis bark water extract (PYBWE) is effective in increasing HA synthesis through several natural product screenings. Therefore, in this study, the effects of PYBWE on HA production in HaCaT cells and the corresponding signaling pathways and tight junction-related protein expression patterns were evaluated.
Experimental
Materials – PYBWE was purchased from Chamhanyak-cho (Gyeongsangbuk-do Yeongcheon-si, Korea). PYBWE was hot air dried at 50oC for 4 h and extracted by repeating the process twice for 4 h at 120oC in a heating mantle (MS-DM607; M-TOPS, Korea) with 200 g of PYBWE and 2,000 mL of distilled water. The extract was filtered through 150 mm filter paper (Advantec, Tokyo, Japan), concentrated under reduced pressure at 47oC using an evaporator (CH-9230; BUCHI Labortechnik AG, Flawil, Switzerland), and lyophilized at -60oC for 24 h. The obtained powder sample extract was 11.2 g.
Cell culture – HaCaT cells were purchased from Cell Line Service (CLS) and cultured at 37oC and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; GenDEPOT, Baker, TX, USA) supplemented with 10% fetal bovine serum (GenDEPOT) and 1% penicillin/streptomycin (GenDEPOT). Cell viability – An MTT (3-(4,5-dimethylthiazol-2-yl)2,5- diphenyltetrazolium bromide) assay was performed to measure the viability of cells treated with PYBWE. HaCaT cells (6 × 103 cells/well) were dispensed into a 96- well plate and incubated for 24 h in an incubator at 37oC and 5% CO2. After the supernatant was removed, PYBWE (25, 50, 100, 200, and 400 µg/mL) was added, and the mixture was allowed to react for 24 h. Then, 5 mg/mL MTT (Bio Base, Canada) was added for 4 h, dimethyl sulfoxide (DMSO; Bio Base) was added and absorbance was measured at 570 nm via a spectrophotometer (SpectraMax 190; Molecular Devices, Sunnyvale, CA, USA). Cell viability is expressed as a percentage relative to that of the control. The average value of the three repeated experiments was obtained, and the experiment was conducted at a nontoxic concentration.
Enzyme-linked immunosorbent assay (ELISA) – To determine the amount of HA produced by HaCaT cells treated with PYBWE, HaCaT cells (2 × 105 cells/well) were dispensed into a 6-well plate and then incubated in an incubator at 37oC and 5% CO2 for 24 h. PYBWE was added at various concentrations (25, 50, and 100 µg/mL) and allowed to react for 24 h. The supernatant was obtained and centrifuged at 4oC and 12,000 rpm for 15 min using a Hyaluronan Enzyme-Linked Immunosorbent Assay Kit (Echelon Biosciences, Salt Lake City, UT, USA). All- trans retinoic acid (ATRA) (1 µM) was used as a positive control.
Immunofluorescence staining – To determine the amount of intracellular HAS-2 secreted by PYBWE- treated HaCaT cells, a sterilized cover glass was placed in a 12-well plate with 2 × 104 cells/well, and the cells were cultured in an incubator for 24 h at 37oC and 5% CO2. The supernatant was removed, the samples were washed three times with PBS, the cover glass was transferred, and PYBWE (25, 50, and 100 µg/mL) was added for 24 h. After reacting in cold 100% methanol for 5 min at room temperature, the samples were washed three times with PBS and then fixed again with 4% paraformaldehyde at room temperature for 10 min. After the cells were lysed with 0.1% to 0.25% Triton X-100, 1% BSA and 22.52 mg/mL glycine in PBST (PBS with 0.1% Tween 20) were added to the cells for blocking for 30 min. Cells were then incubated overnight with the primary antibody at 4oC. After the samples were washed three times with PBS, they were incubated with 1% BSA and the secondary antibody for 3 h without being exposed to light. They were then washed three times, and mounting solution (with DAPI) (ab104139; Abcam, Cambridge, UK) was added to the glass slides for fluorescence observation. ATRA (1 µM) was used as a positive control.
Quantitative real-time polymerase chain reaction (qRT-PCR) – To determine whether PYBWE increases the mRNA expression levels of skin barrier- and moisture- related genes, HaCaT cells (6 × 105 cells/well) were dispensed into 60 mm plates and incubated in an incubator at 37oC for 24 h at 5% CO2. The supernatant was removed. Then, PYBWE (25, 50, and 100 µg/mL) was added, using ATRA (1 µM) as a positive control, and reacted for 24 h. After the reaction, RNA was extracted using TRIzol (Ambion, Waltham, MA, USA), dissolved in diethylpyrocarbonate (DEPC)-treated water (Sigma, St. Louis, MO, USA), and quantified. The quantified RNA was reverse transcribed using a cDNA synthesis kit (Revertra ACE-A-; Toyobo, Osaka, Japan). To determine the expression levels of genes related to the skin barrier and moisture, cDNA, TaqMan Master Mix (TaqMan, Foster City, CA, USA), DEPC, and primers were mixed, and qRT‒PCR was performed. The probes used in the experiment are shown in Table 1.
Western blot analysis – HaCaT cells (6 × 105 cells/well) were dispensed into 60 mm plates and incubated in an incubator for 24 h at 37oC and 5% CO2. Thereafter, the supernatant was removed, and PYBWE (25, 50, and 100 µg/mL) and the positive control ATRA (1 μM) were added and allowed to react for 15 min. After washing once with PBS, the supernatant was removed, and the cells were lysed with RIPA lysis extraction buffer (Thermo Fisher, Waltham, MA, USA). Proteins obtained by centrifugation at 4oC and 12,000 rpm for 20 min were quantified using a BCA Protein Assay Kit (Thermo Fisher Scientific). The quantified proteins were stained with 5 × SDS‒PAGE loading dye (Binoseang, Korea), loaded onto a 10% polyacrylamide gel, and separated by electrophoresis. The membrane was transferred, blocked with 5% skim milk for 1 h, and incubated overnight with primary antibodies. Protein expression was confirmed by washing three times with Tris-buffered saline with Tween 20 (TBST) and washing again after 3 h of treatment with the secondary antibody. The antibodies used in the experiment are shown in Table 2.
High-performance liquid chromatography – Naringenin was quantified using a SHIMADZU LC-2040C 3D System and a Eurosper 100-5 C18 column (250 × 3 mm). The mobile phases were acetonitrile (A) and water (B), the flow rate of the mobile phase was set to 0.8 mL/min, and the column oven temperature was set to 25oC. The UV wavelength was set to 280 nm, and the injection volume was 10 μL. PYBWE was used at a concentration of 10 mg/mL, and naringenin was used after preparing a standard stock solution of 500 µg/mL and diluting it to concentrations of 10, 62.5, 125, 250, and 500 µg/mL. The solvents were acetonitrile for high-performance liquid chromatography (HPLC) (Daejung Chemicals & Metals) and water for HPLC (J.T. Baker). Naringenin, an indicator component of PYBWE, was purchased from Sigma Aldrich. The conditions for the naringenin analysis are shown in Table 3.
Statistical analysis – All experiments were repeated three times and results are expressed as the means ± standard errors. To determine the effect of PYBWE, comparisons between groups were performed using Student’s t-tests. The statistical significance level was p < 0.05.
Results and Discussion
P.yedoensis Matsumura is a deciduous tree belonging to the rose family and has been used in daily life for various practical purposes.24,25P. yedoensis contains various flavonoids, such as naringenin, sakuranetin, and prunetin, of which the content of naringenin is the highest.26 Royal cherry trees have various physiological effects, such as vascular relaxation, anti-inflammatory, and wound healing effects.27,28 However, studies on HA synthesis and tight junction-related proteins are still insufficient; therefore, in this study, the effects of PYBWE on the expression levels and mechanisms of action of mRNAs and proteins related to HA and tight junctions were evaluated.
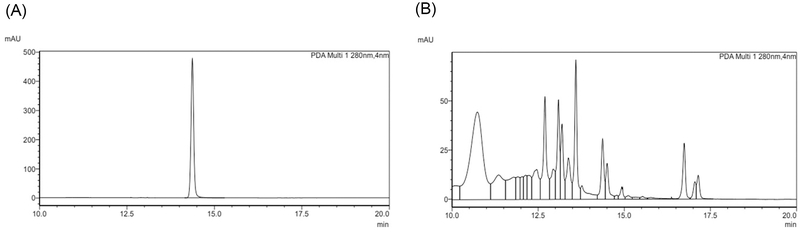
HPLC was performed to analyze the content of naringenin, an indicator component in PYBWE. A calibration curve was created where the y-axis represents the peak area and the x-axis represents the concentration (ppm) of the naringenin solution. The correlation coefficient (R2) for the prepared calibration curve (y = ax − b) was 0.997. PYBWE and naringenin were detected at 14.372 min and 14.371 min, respectively. In PYBWE, the content of naringenin was 12.14 µg/mL (Fig. 1).
The evaporation of moisture in the skin causes itching, redness, scales, and other symptoms and allows external substances to pass through the skin more easily.29,30 HA is a natural moisturizing agent that maintains the skin barrier by preventing the evaporation of moisture in the epidermis; it is also involved in physiological functions, such as the storage and diffusion of nutrients and cell migration.31,32 A decrease in HA in the skin causes damage to the skin barrier, which facilitates the penetration of external substances, resulting in skin diseases, such as atopy and psoriasis. Therefore, substances that promote the synthesis of HA or inhibit decomposition are effective in maintaining skin moisture and improving structural stability.33 Recently, increasing HAS gene expression to increase HA production has been evaluated.34 A recent study has shown that the treatment of skin keratinocytes with HA improves skin barrier function by increasing tight junction protein levels.17
All-trans retinoic acid (ATRA), used as a positive control in experiments, is widely used for controlling cell growth and differentiation and for treating skin diseases; it induces HAS-2 mRNA expression in HaCaT cells.35,36
In this study, the effects of PYBWE on HA synthesis and tight junction function in An MTT assay was performed to evaluate the cytotoxicity of PYBWE (25, 50, 100, 200, and 400 µg/mL) in HaCaT cells. Setting viability in the control to 100%, PYBWE treatment led to viabilities of 99.34 ± 10.97%, 95.90 ± 9.31%, 87.74 ± 6.82%, 76.86 ± 6.15%, and 73.22 ± 4.04%, indicating that PYBWE was toxic at 200 µg/mL or greater. Subsequent analyses were conducted with PYBWE at concentrations of 25, 50, and 100 µg/mL (Fig. 2). PYBWE was used at a concentration of 10 mg/mL after dissolution in serum-free DMEM.
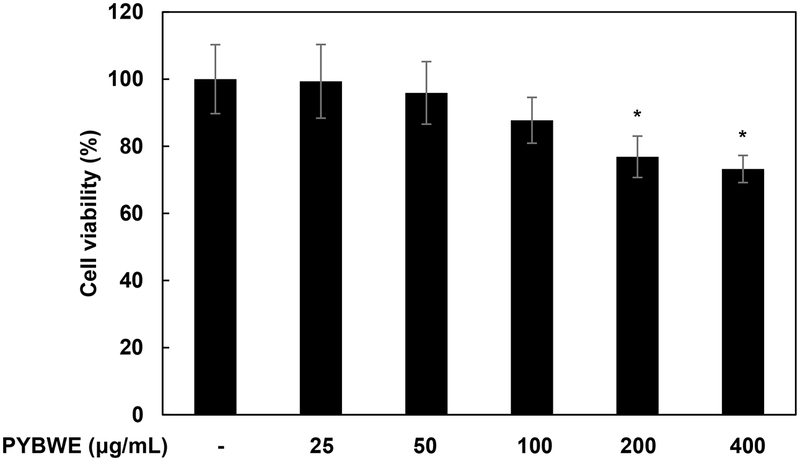
Effects of PYBWE on HaCaT cell viability. Cells were treated with different concentrations of PYBWE (25, 50, 100, 200, and 400 μg/mL) for 24 h. Cell viability was measured using the MTT assay, as described in the Materials and Methods section. The results are expressed as the means ± SDs of three independent experiments. *p < 0.05 compared with the control.
ELISA was used to quantify HA production induced by PYBWE in HaCaT cells. Treatment with PYBWE at concentrations of 25, 50, and 100 µg/mL increased the amount of HA produced in a concentration-dependent manner to 9.68%, 47.86%, and 139.05% respectively. In the positive control group (treated with ATRA), HA production also increased (Fig. 3).
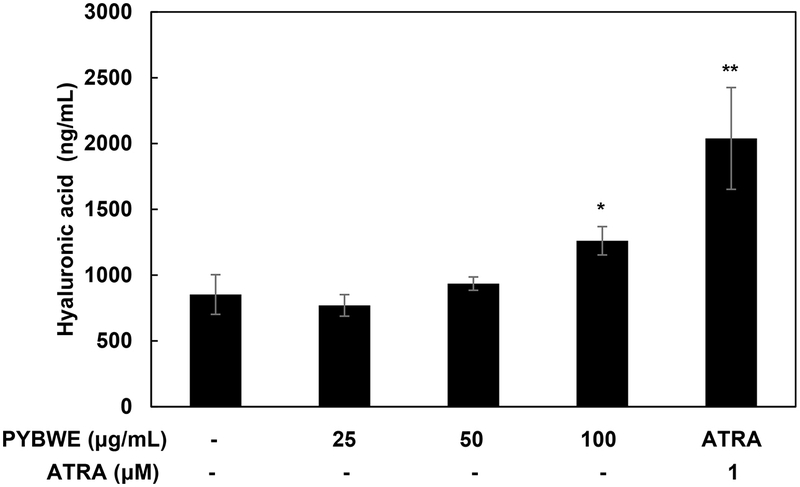
Effects of PYBWE on HA production in HaCaT cells. The cells were treated with different concentrations of PYBWE (25, 50, and 100 μg/mL) for 24 h. ATRA (1 μM) was used as a positive control. The results are expressed as the means ± SDs of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 compared with the control.
HA is synthesized by HAS, a synthase that can be divided into three forms: HAS-1, HAS-2, and HAS-3. The latter two (HAS-2 and HAS-3) play important roles in determining the length and structure of HA. However, HAS-1 has a relatively weak ability to stably maintain the HA chain in the cell membrane; thus, the conservation of HA is limited.8,37
The HAS-2 protein expression level in PYBWE-treated HaCaT cells was confirmed by immunofluorescence staining. HAS-2 was expressed after treatment with PYBWE at all concentrations (25, 50, and 100 g/mL) and after treatment with ATRA, a positive control (Fig. 4). HaCaT cells were treated with PYBWE to evaluate the mRNA expression levels of HAS-2 and HAS-3 through qRT‒PCR. When PYBWE was added at concentrations of 25, 50, and 100 μg/mL, HAS-2 mRNA expression levels increased in a concentration-dependent manner by 1.41 ± 0.67, 1.67 ± 0.32, and 3.96 ± 1.77 times, whereas HAS-3 mRNA expression levels decreased in a concentration-dependent manner by 0.71 ± 0.16, 0.44 ± 0.13, and 0.25 ± 0.07 times. ATRA clearly increased HAS-2 and HAS-3 levels (Fig. 5). These results show a similar trend with the increase in HAS-2 mRNA after EGF treatment. In this study, there were no changes in the expression of HAS-1 and HAS-3, but it was reported that the increased expression of HAS- 2 promoted HA synthesis. This result provides important implications that HAS-2 acts as a major regulator of HA synthesis.48 Our experimental results also confirmed that the increased expression of HAS-2 is closely related to HA synthesis. Thus, HA synthesis is mediated mainly by HAS-2, emphasizing the importance of HAS-2 in the control of skin moisture.
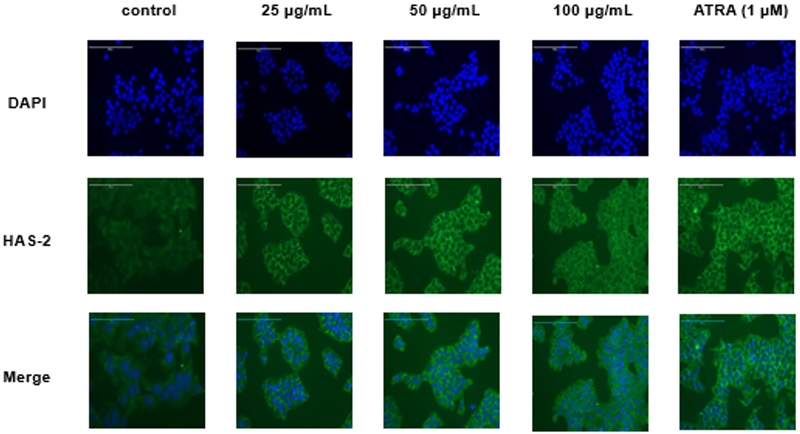
Effects of PYBWE on HAS-2 protein expression in HaCaT cells. The cells were treated with different concentrations of PYBWE (25, 50, and 100 μg/mL) for 24 h. ATRA (1 μM) was used as a positive control.
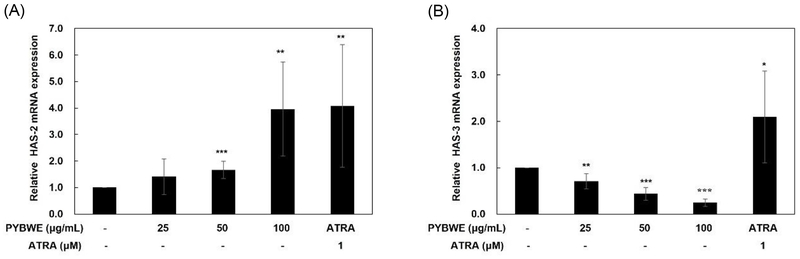
Effects of PYBWE on HAS-2 and HAS-3 mRNA levels in HaCaT cells. The cells were treated with different concentrations of PYBWE (25, 50, and 100 μg/mL) for 24 h. ATRA (1 μM) was used as a positive control. The results are expressed as the means ± SDs of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 versus the control.
Since most cell surface receptors use one or more mitogen-activated protein kinase (MAPK) pathways, this study focused on the MAPK pathway leading to activation.38 HaCaT cells were treated with PYBWE, and western blotting was performed to determine whether the increase in HAS-2 mRNA expression influences MAPK signaling pathway protein phosphorylation. Compared with levels in the control, the protein expression levels of ERK, JNK, and p38 increased in a PYBWE concentration-dependent manner (Fig. 6A–D). Cyclical AMP response element- binding protein (CREB) is a transcription factor that is regulated by ERK; it is important for various cellular processes, including proliferation, differentiation, and adaptation. HAS-2 expression is dependent on ERK and CREB.39 Thus, the phosphorylation of CREB is expected to increase through PYBWE treatment in HaCaT cells; thus, HA synthesis is mediated by the MAPK and CREB pathways. In this study, CREB protein expression levels also increased in response to PYBWE in a concentration- dependent manner (Fig. 6E–F). The regulation of HAS expression through the Akt and NF-κB signaling pathways is clearly established.40 As there is evidence that HA synthesis involves HAS-2 expression via the AKT and NF-κB signaling pathways, these signaling pathways were also evaluated through western blotting using HaCaT cells with PYBWE. AKT and NF-κB increased in a concentration-dependent manner (Fig. 6G–J). In addition, the role of the MAPK signaling pathway in PYBWE- induced changes in HAS-2 expression was evaluated. In response to treatment with the ERK inhibitor U016, p38 inhibitor SB203580, or JNK inhibitor SP600125, HAS-2 expression was lower than that in the group treated with PYBWE alone (Fig. 7). These findings confirmed that PYBWE regulates HAS-2 expression through the MAPK, CREB, Akt, and NF-κB signaling pathways.
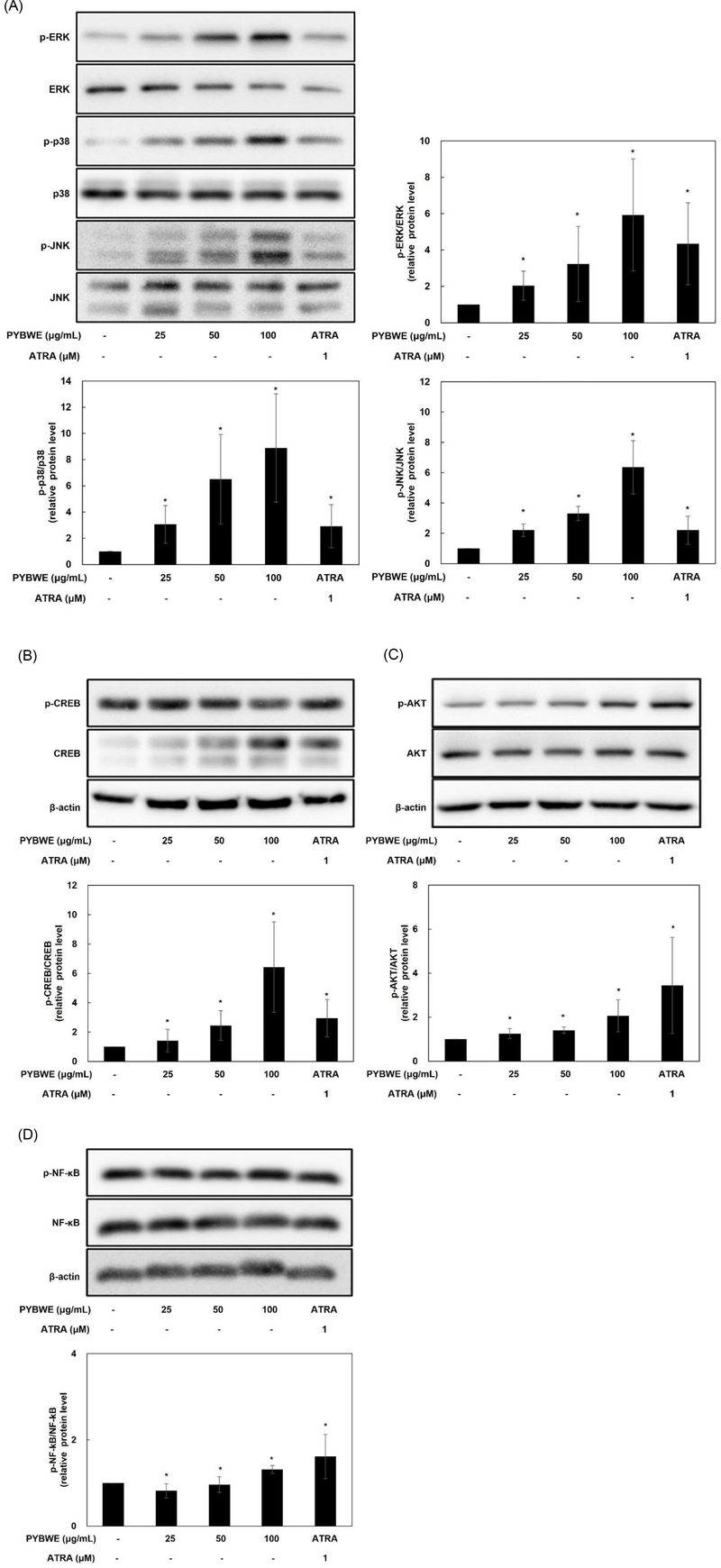
Effects of PYBWE on MAPK, CREB, Akt, and NF-κB protein expression in HaCaT cells. The cells were treated with different concentrations of PYBWE (25, 50, and 100 μg/mL) for 24 h. ATRA (1 μM) was used as a positive control. (A–D) PYBWE increased the phosphorylation of MAPK, CREB, Akt, and NF-κB. Data were analyzed using ImageJ. The results are expressed as the means ± SDs of three independent experiments. *p < 0.05 compared with the control.
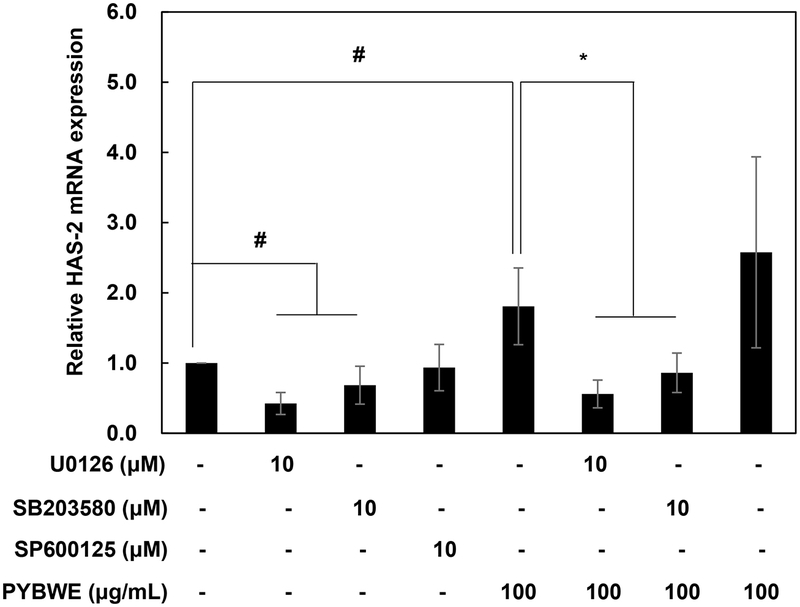
Inhibitory effects of the MAPK signaling pathway on PYBWE-induced HAS-2 expression in HaCaT cells. The cells were treated with different concentrations of PYBWE (25, 50, and 100 μg/mL) for 24 h. U0126 is an ERK inhibitor. SB2023580 is a p38 inhibitor. SP300125 is a JNK inhibitor. The results are expressed as the means ± SDs of three independent experiments. #p < 0.05 compared with the control. *p < 0.05 compared with the PYBWE.
The epidermis of the skin plays an important barrier function, particularly the outmost layer, the stratum corneum. The normal skin barrier plays an important role in maintaining the health of the skin and preventing damage from the external environment.41
The tight junctions are located in the stratum granulosum, the middle of the three layers.42 Damage to tight junctions causes the destruction of the stratum corneum, thereby increasing the mobility of external substances. This may affect the differentiation of keratinocytes and consequently lead to percutaneous water loss.43 Tight junctions are composed of three main proteins: claudins, occludins, and functional adhesion molecules (JAM). They play an important role in forming a hydrophilic space through the cell membrane. In particular, claudin accounts for the largest proportion of these proteins and is expressed in all epithelial tissues.44 There are 23 known claudin types, of which deficiencies in CLDN-1 and CLDN-4 may cause a decrease in skin barrier function, thereby increasing the likelihood of developing inflammatory diseases, such as atopy.14 These tight junction proteins interact with tight junction cytoplasmic proteins, such as ZO-1, ZO-2, ZO-3 multiple PDZ domain protein (MUPP)-1, and PALS1-associated tight junction protein (PATJ), to maintain and stabilize tight junctions.45 HaCaT cells were treated with PYBWE for analyses of the mRNA expression levels of CLDN-1, CLDN-3, CLDN-4, and F11r using qRT‒PCR. In particular, after treatment with PYBWE at concentrations of 25, 50, and 100 µg/mL, CLDN-1 levels did not differ significantly from those in the control group (1.20 ± 0.48, 0.77 ± 0.32, and 1.00 ± 0.43, respectively). Additionally, CLDN-3 levels did not differ significantly (1.44 ± 0.23, 1.18 ± 1.13, and 1.46 ± 0.33, respectively). The expression levels of CLDN-4 were 0.97 ± 0.23, 1.13 ± 0.33, and 1.72 ± 0.38 times higher in the PYBWE treatment groups than in the control group, indicating that the expression level increased only at a concentration of 100 µg/mL PYBWE. F11r levels were reduced in a concentration-dependent manner to 0.85 ± 0.3, 0.59 ± 0.43, and 0.45 ± 0.28 (Fig. 8). In the positive control group, CLDN-1 expression was lower than that in the other groups, consistent with previous results showing that ATRA inhibits the activity of CLDN-1.46 Western blotting confirmed that ATRA treatment resulted in higher CLDN-4 phosphorylation in HaCaT cells were treated with PYBWE than that in the control (Fig. 9). According to the results of previous studies, claudin recruits occludin to tight junctions, and a decrease in occludin does not affect the formation of tight junctions. In addition, tight junctions can be formed in fibroblasts without tight junctions via claudin.47 These results suggest that claudin plays a central role and occludin plays an auxiliary role in the formation of tight junctions. The results of this study demonstrated that HA is synthesized via HAS-2 expression in HaCaT cells treated with PYBWE, and these effects on HA synthesis are mediated by CLDN-4, thereby improving tight junction function and enhancing the moisture barrier of the skin, with the potential to improve skin diseases.
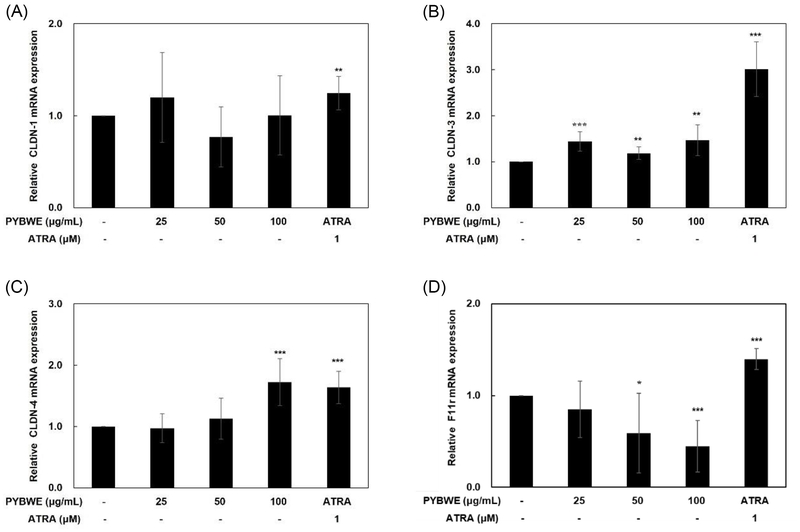
Effects of PYBWE extract on CLDN-1, CLDN-3, CLDN-4, and F11r mRNA levels in HaCaT cells. HaCaT cells were treated with different concentrations of PYBWE (25, 50, and 100 μg/mL) for 24 h. ATRA (1 μM) was used as a positive control. The results are expressed as the means ± SDs of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 compared with the control.
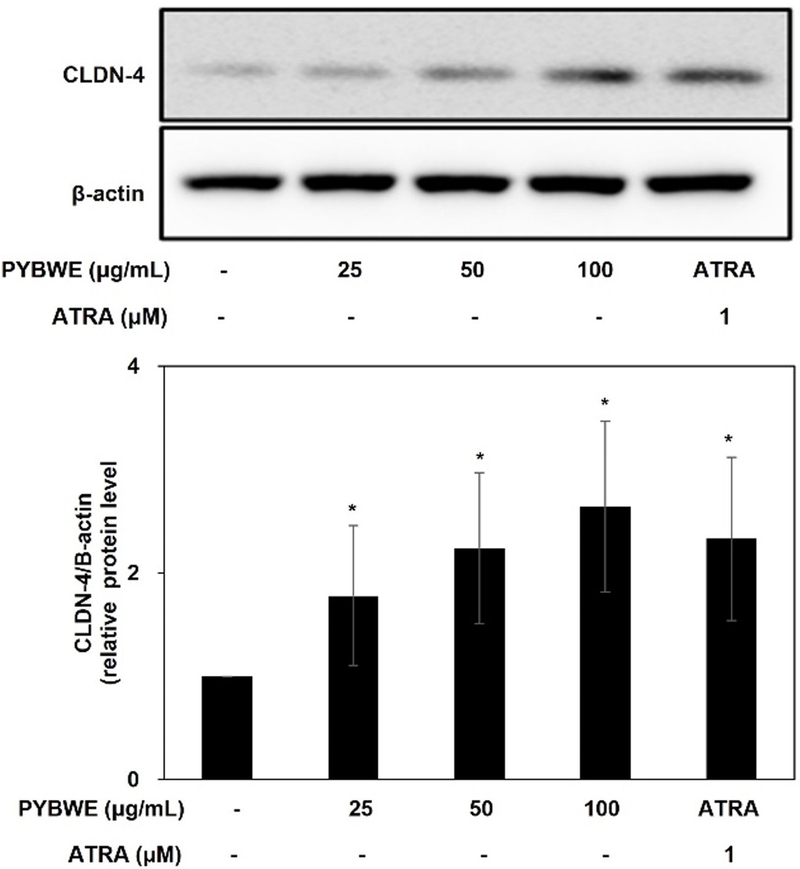
Effects of PYBWE on CLDN-4 protein expression in HaCaT cells. The cells were treated with different concentrations (25, 50, and 100 μg/mL) for 24 h. ATRA (1 μM) was used as a positive control. PYBWE increased the phosphorylation of CLDN-4. The data were analyzed using ImageJ, and the results are expressed as the means ± SDs of three independent experiments. *p < 0.05 compared with the control.
Acknowledgments
Conflicts of Interest
The authors declare that they have no conflicts of interest.
REFERENCES
-
Biggs, L. C.; Kim, C. S.; Miroshnikova, Y. A.; Wickström, S. A. J. Invest. Dermatol. 2020, 140, 284–290.
[https://doi.org/10.1016/j.jid.2019.06.137]

-
Chambers, E. S.; Vukmanovic‐Stejic, M. Immunology 2020, 160, 16–125.
[https://doi.org/10.1111/imm.13152]

-
D’Orazio, J.; Jarrett, S.; Amaro-Ortiz, A.; Scott, T. Int. J. Mol. Sci. 013, 14, 12222–12248.
[https://doi.org/10.3390/ijms140612222]

-
Lefèvre-Utile, A.; Braun, C.; Haftek, M.; Aubin, F. Int. J. Mol. Sci. 2021, 22, 11676.
[https://doi.org/10.3390/ijms222111676]

-
Baek, J. H.; Lee, S. E.; Choi, K. J.; Choi, E. H.; Lee, S. H. Yonsei Med. J. 2013, 54, 523–528.
[https://doi.org/10.3349/ymj.2013.54.2.523]

-
Juncan, A. M.; Moisă, D. G.; Santini, A.; Morgovan, C.; Rus, L.-L.; Vonica-Țincu, A. L.; Loghin, F. Molecules 2021, 26, 4429.
[https://doi.org/10.3390/molecules26154429]

-
Marinho, A.; Nunes, C.; Reis, S. Biomolecules 2021, 11, 1518.
[https://doi.org/10.3390/biom11101518]

-
Volpi, N.; Schiller, J.; Stern, R.; Soltés, L. Curr. Med. Chem. 2009, 16, 1718–1745.
[https://doi.org/10.2174/092986709788186138]

-
Hon, K. L.; Leung, A. K. C.; Barankin, B. Am. J. Clin. Dermatol. 2013, 14, 389–399.
[https://doi.org/10.1007/s40257-013-0033-9]

-
Lee, J.-O.; Hwang, S.-H.; Shen, T.; Kim, J. H.; You, L.; Hu, W.; Cho, J. Y. J. Ginseng Res. 2021, 45, 354–360.
[https://doi.org/10.1016/j.jgr.2020.12.003]

-
Adil, M. S.; Narayanan, S. P.; Somanath, P. R. Tissue Barriers 2021, 9, 1848212.
[https://doi.org/10.1080/21688370.2020.1848212]

-
Proksch, E.; Brandner, J. M.; Jensen, J.‐M. Exp. Dermatol. 2008, 17, 1063–1072.
[https://doi.org/10.1111/j.1600-0625.2008.00786.x]

-
González-Mariscal , L.; Betanzos, A.; Nava, P.; Jaramillo, B. E. Prog. Biophys. Mol. Biol. 2003, 81, 1–44.
[https://doi.org/10.1016/S0079-6107(02)00037-8]

-
Findley, M. K.; Koval, M. IUBMB Life 2009, 61, 431–437.
[https://doi.org/10.1002/iub.175]

-
Kirschner, N.; Bohner, C.; Rachow, S.; Brandner, J. M. Arch. Dermatol. Res. 2010, 302, 483–493.
[https://doi.org/10.1007/s00403-010-1058-z]

-
Katsarou, S.; Makris, M.; Vakirlis, E.; Gregoriou, S. J. Clin. Med. 2023, 12, 1538.
[https://doi.org/10.3390/jcm12041538]

-
Tokumasu, R.; Yamaga, K.; Yamazaki, Y.; Murota, H.; Suzuki, K.; Tamura, A.; Bando, K.; Furuta, Y.; Katayama, S.; Tsukita, S. Proc. Natl. Acad. Sci. U S A 2016, 113, E4061–E4068.
[https://doi.org/10.1073/pnas.1525474113]

-
Park, H.-Y.; Kweon, D.-K.; Kim, J.-K. Bioact. Carbohydr. Diet. Fibre 2023, 30, 100374.
[https://doi.org/10.1016/j.bcdf.2023.100374]

-
Rauhala, L.; Jokela, T.; Kärnä, R.; Bart, G.; Takabe, P.; Oikari, S.; Tammi, M. I.; Pasonen-Seppänen, Sanna; Tammi, R. H. Biochem. J. 2018, 475, 1755–1772.
[https://doi.org/10.1042/BCJ20180054]

-
Ohno, S.; Im, H.-J.; Knudson, C. B.; Knudson, W. J. Biol. Chem. 2006, 281, 17952–17960.
[https://doi.org/10.1074/jbc.M602750200]

-
Li, L.; Asteriou, T.; Bernert, B.; Heldin, C.-H.; Heldin, P. Biochem. J. 2007, 404, 327–336.
[https://doi.org/10.1042/BJ20061757]

-
Lee, K.; Ham, I.; Yang, G.; Lee, M.; Bu, Y.; Kim, H.; Choi, H.-Y. BMC Complement. Altern. Med. 2013, 13, 31.
[https://doi.org/10.1186/1472-6882-13-31]

-
Lee, J.; Yang, G.; Lee, K.; Lee, M.-H.; Eom, J.-W.; Ham, I.; Choi, H.-Y. BMC Complement. Altern. Med. 2013, 13, 92.
[https://doi.org/10.1186/1472-6882-13-92]

-
Velmurugan, P.; Cho, M.; Lim, S.-S.; Seo, S.-K.; Myung, H.; Bang, K.-S.; Oh, B.-T. Mater. Lett. 2015, 138, 272–275.
[https://doi.org/10.1016/j.matlet.2014.09.136]

- Kim, E.-N.; Kim, S.-C.; Jeong, G.-S. Kor. J. Pharmacogn. 2019, 50, 226–231.
-
Wang, Y.; Li, W.; Xu, S.; Hu, R.; Zeng, Q.; Liu, Q.; Li, S.; Lee, H.; Chang, M.; Guan, L. Cytotechnology 2019, 71, 475–487.
[https://doi.org/10.1007/s10616-018-0215-7]

-
Lee, J.-H.; Lee, K.; Lee, M.-H.; Kim, B.; Chinannai, K. S.; Hur, H.; Ham, I.; Choi, H.-Y. Evid. Based Complement. Alternat. Med. 2017, 1, 7812598.
[https://doi.org/10.1155/2017/7812598]

- Tončić, R. J.; Kezić, S.; Hadžavdić, S. L.; Marinović, B. Clin. Dermatol. 2018, 36, 109–115.
-
Bouwstra, J. A.; Dubbelaar, F. E. R.; Gooris, G. S.; Ponec, M. Acta Derm. Venereol. Suppl. 2000, 208, 23–30.
[https://doi.org/10.1080/000155500750042826]

-
Dübe, B.; Lüke, H. J.; Aumailley, M.; Prehm, P. Biochim. Biophys. Acta 2001, 1538, 283–289.
[https://doi.org/10.1016/S0167-4889(01)00079-9]

-
Juncan, A. M.; Moisă, D. G.; Santini, A.; Morgovan, C.; Rus, L.- L.; Vonica-Țincu, A. L.; Loghin, F. Molecules 2021, 26, 4429.
[https://doi.org/10.3390/molecules26154429]

-
Lee, Y. I.; Lee, S. G.; Kim, J.; Choi, S.; Jung, I.; Lee, J. H. Int. J. Mol. Sci. 2021, 22, 10189.
[https://doi.org/10.3390/ijms221910189]

-
Kim, N. Y.; Won, K. J.; Kim, D. Y.; Lee, D. K.; Kim, Y. Y.; Lee, H. M. Heliyon 2024, 10, e24434.
[https://doi.org/10.1016/j.heliyon.2024.e24434]

-
Trinh, T.-T. T.; Choi, J. H.; Yang, J.-E.; Kim, W. H.; Chien, P. N.; Le, L. T. T.; Ngan-Giang, N.; Nga, P. T.; Nam, S.-Y.; Heo, C.-Y. Skin Res. Technol. 2024, 30, e13682.
[https://doi.org/10.1111/srt.13682]

-
Szymański, Ł.; Skopek, R.; Palusińska, M.; Schenk, T.; Stengel, S.; Lewicki, S.; Zelent, A. Cells 2020, 9, 2660.
[https://doi.org/10.3390/cells9122660]

-
Saavalainen, K.; Pasonen-Seppänen, S.; Dunlop, T. W.; Tammi, R.; Tammi, M. I.; Carlberg, C. J. Biol. Chem. 2005, 280, 14636–14644.
[https://doi.org/10.1074/jbc.M500206200]

-
Siiskonen, H.; Pasonen-Seppänen, S.; Rilla, K. Front. Immunol. 2015, 6, 43.
[https://doi.org/10.3389/fimmu.2015.00043]

-
Pearson, G.; Robinson, F.; Beers Gibson, T.; Xu, B. E.; Karandikar, M.; Berman, K.; Cobb, M. H. Endocr. Rev. 2001, 22, 153–183.
[https://doi.org/10.1210/edrv.22.2.0428]

-
Shaywitz, A. J.; Greenberg, M. E. Annu. Rev. Biochem. 1999, 68, 821–861.
[https://doi.org/10.1146/annurev.biochem.68.1.821]

-
Kim, S. J.; Kwon, M. S.; Oh, S. R.; Jeon, S. H.; Lee, P. J.; Park, S. K.; Kim, T. J.; Kim, Y. M. Biotechnol. Bioprocess Eng. 2022, 27, 51–60.
[https://doi.org/10.1007/s12257-020-0341-x]

-
Elias, P. M. J. Invest. Dermatol. 2005, 125, 183–200.
[https://doi.org/10.1111/j.0022-202X.2005.23668.x]

-
Yoshida, K.; Yokouchi, M.; Nagao, K.; Ishii, K.; Amagai, M.; Kubo, A. J. Dermatol. Sci. 2013, 71, 89–99.
[https://doi.org/10.1016/j.jdermsci.2013.04.021]

-
Sugawara, T.; Iwamoto, N.; Akashi, M.; Kojima, T.; Hisatsune, J.; Sugai, M.; Furuse, M. J. Dermatol. Sci. 2013, 70, 12–18.
[https://doi.org/10.1016/j.jdermsci.2013.01.002]

-
Citi, S.; Cordenonsi, M. Biochim. Biophys. Acta 1998, 1448, 1–11.
[https://doi.org/10.1016/S0167-4889(98)00125-6]

-
Heinemann, U.; Schuetz, A. Int. J. Mol. Sci. 2019, 20, 6020.
[https://doi.org/10.3390/ijms20236020]

- Li, J.; Li, Q.; Geng, S. Int. J. Mol. Med. 2019, 43, 1789–1805.
-
Furuse, M.; Sasaki, H.; Fujimoto, K.; Tsukita, S. J. Cell Biol. 1998, 143, 391–401.
[https://doi.org/10.1083/jcb.143.2.391]

-
Pienimäki, J. P.; Rilla, K.; Fülöp, C.; Sironen, R. K.; Karvinen, S.; Pasonen, S.; Lammi, M. J.; Tammi, R.; Hascall, V. C.; Tammi, M. I. J. Biol. Chem. 2001, 276, 20428–20435.
[https://doi.org/10.1074/jbc.M007601200]