Cytotoxic Compounds Obtained through Cell Viability Screening of Fungal Extracts, Isolated from Urban Soil Samples I
Abstract
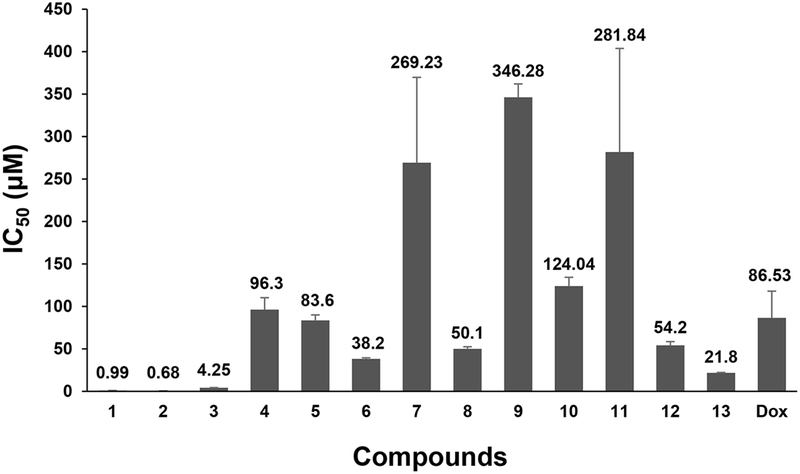
Soil microorganisms have been reported to interact with plants, playing key roles such as providing nutrients essential for plant growth and protecting them from plant pathogens. Additionally, various bioactive molecules from soil significantly contribute to controlling diseases threatening human such as cancer and infectious diseases. Considering the crucial roles and potential of these soil microorganisms, the Natural Products Drug Discovery research group (NPDD) at Duksung Women’s University collected soil samples from various environments, isolated fungi from the soil, and aimed to discover bioactive compounds. In this process, 96 fungal strains were cultured on small scale to generate their ethyl acetate extracts, which were subsequently screened for cell viability using MDA-MB-231 triple-negative breast cancer cells. Among them, a subset of 17 with over 60% inhibitory activity were selected as top-performing strains, and other 13 strains with below 60% were chosen for large-scale fermentation candidates by HR-ESI-MS dereplication process to discover new bioactive molecules. Chemical investigation of a part of these large-scale fermentation candidates led to the isolation of 13 major metabolites (1–13), and all isolates were evaluated for their cytotoxicity against MDA-MB-231 cells. As a result, strigaibol C (1), trichoguizaibol J (2), beauvericin (3), and sclerotioramine (13) showed strong inhibitory activities with IC50 values of 0.99, 0.68, 4.25, and 21.8 μM, respectively.
Keywords:
Soil fungi, Cytotoxicity, MDA-MB-231, Peptaibols, Penicillium, TrichodermaIntroduction
Natural products grown by absorbing nutrients from the soil provide essential resources for human life, not only food such as fruits and vegetables but also materials for clothing and textiles.1,2 Moreover, well-known natural medicines such as acetylsalicylic acid, morphine and paclitaxel are derived from plants.3–5 Also, compounds obtained from microbes such as penicillin and streptomycin have become indispensable drugs for humanity, which have been used as antibiotics.6,7 Even recently, ivermectin and its analogues, isolated from soil-derived actinomycetes, have been continuously used as an antiparasitic drug.8 In addition to the above facts, the ongoing research about the interactions between soil microorganisms and medicinal plants has led us to focus on the metabolites of soil fungi.9 Furthermore, these soil microorganisms exhibit different distributions according to natural environments, and since even large urban areas can possess sufficiently diverse natural environments, it is possible to obtain a variety of soil fungi.10 Based on this trend, the Natural Products Drug Discovery research group (NPDD) at Duksung Women’s University aimed to discover cytotoxic compounds through cell viability screening using the MDA-MB-231 triplenegative breast cancer cell line. First, we began to isolate fungi from soil samples collected from the territory of inside or near Duksung Women’s University, resulting in the identification of 96 fungal strains from 14 different soil samples. These fungal strains were cultured on small scale on Cheerios oat cereal, and their extracts were used for bioactivity screening.11–14 From the fungi that showed great IC50 values against MDA-MB-231 cells, their major compounds were isolated and further evaluated for their cytotoxicity. Herein, we describe the process of finding cytotoxic metabolites from soil samples, and isolation, purification, and cytotoxic activity evaluation of major components produced by fungal strains collected from inside or near Duksung Women’s University.
Experimental
General experimental procedures – Nuclear magnetic resonance (NMR) spectra were obtained on Varian NMR spectrometers (400 MHz for 1H and 100 MHz for 13C). The preparative HPLC was performed using Waters HPLC system equipped with pumps, a 996 photodiode-array detector, and a Luna 5 μm C18 column (100 Å, 250 × 21.2 mm) with a flow rate of 10 mL/min. The semi-preparative HPLC were conducted on the same Waters HPLC system using a Luna 5 μm C18 (100 Å, 250 × 10 mm) with a flow rate of 4 mL/min. The HPLC analysis was carried out on the same Waters HPLC system using a Luna 5 μm C18 (100 Å, 250 × 4.6 mm) with a flow rate of 1 mL/min. HRESIMS analysis was performed using a UHPLC-HROrbitrap mass spectrometer (Thermo Fisher Scientific) system equipped with a diode array detector and an YMCTriart C18 column (2.0 μm, 100 × 2.1 mm, i.d., flow rate 0.3 mL/min). All solvents used were of ACS grade or better.
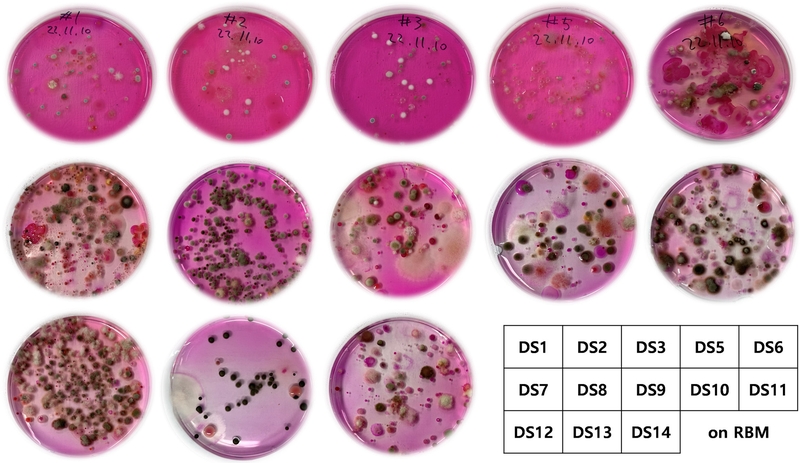
Fungal isolates and fermentation – Fungal strains were isolated from soil samples, which were collected from the territory of inside or near Duksung Women’s University, Seoul, South Korea in November 2022 (DSWU collection, Table 1). Fungi were identified based on the ribosomal internal transcribed spacer (ITS) region (Macrogen Inc. and Biofact Inc.). All fungi were identified by comparing the resulting sequence data with fungal sequences in GenBank. All sequence data were deposited in GenBank (Table 2).
To carry out small-scale cultivation for the cell cytotoxicity screening, fungi were recovered from cryogenic storage (stored in a vial at −80oC as mycelium with 20% glycerol aqueous solution). Following recovery on rose bengal medium plates (15 g agar, 10 g malt extract, 1 g yeast, 0.05 g chloramphenicol, 0.025 g rose bengal, 1 L deionized H2O), fungal mycelia were aseptically cut into small pieces (~ 0.5 cm2) for the small-scale fermentation. Small-scale fermentation was performed using a 250 mL flask with bilayers of Cheerios breakfast cereal as the medium, supplemented with a 0.3% sucrose solution containing 0.005% chloramphenicol. Pieces of mycelia (~ 0.5 cm2) were aseptically inoculated into a 250 flask and the cultures were grown at room temperature for two weeks. Largescale cultivation was also carried out using ten 1 L flasks for the isolation and purification of bioactive compounds in the same method above.
Initial HPLC analysis and dereplication using HRESI-MS – All fungal extracts were analyzed using our inhouse HPLC method [C18, gradient elution with 10 to 100% MeCN in H2O, over 40 min and isocratic 100% MeCN over 10 min using a 1 mL/min flow rate]. The HRESIMS-dereplication process was carried out by comparing the chemical formulas, UV absorbance, and retention times of major peaks in each extract with webbased platform libraries such as GNPS and NPAtlas, as well as our in-house extracts and compounds library.
Extraction and isolation of fungal extracts – All fungal cultures were extracted with EtOAc (large-scale: 0.5 L × 3; small scale: 150 mL × 3) at room temperature overnight. The extract was filtered and evaporated under reduced pressure using a rotary vacuum evaporator to obtain the EtOAc-soluble residue. The EtOAc soluble materials of large-scale cultivation (fraction A) were subjected to silica gel vacuum column chromatography with elution performed using dichloromethane (fraction B), dichloromethane-MeOH (10:1) (fraction C), and MeOH (fraction D). Fraction C was also further fractionated by HP20ss gel vacuum column chromatography into five sample: fractions E (30% MeOH), F (50% MeOH), G (70% MeOH), H (90% MeOH) and I (100% MeOH). Those fractions E-I obtained from large-scale fermentation were isolated and purified using preparative or semi-preparative HPLC. Additionally, the EtOAc extract from small-scale cultivation was directly subjected to preparative or semipreparative HPLC for the purification of major metabolites without vacuum liquid column chromatography process.
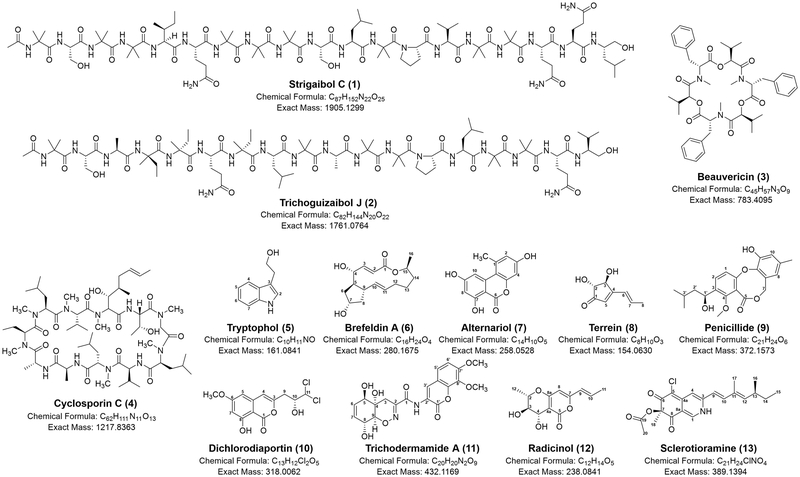
Strigaibol C (1) – Compound 1 was isolated from a large-scale fungal extract of DS7-1 (Trichoderma strigosum). For detailed information on the purification process and structure elucidation, see ref 15.15
Trichoguizaibol J (2) – Metabolite 2 was purified from a large-scale fungal extract of DS9-1 (Trichoderma guizhouense). For detailed information on the purification process and structure elucidation, see ref 16.16
Beauvericin (3) – Small-scale fungal extract of DS1-6 (Fusarium oxysporum, 20 mg) was isolated by semipreparative HPLC [C18, isocratic MeCN-H2O (85:15), flow rate: 4 mL/min] to yield compound 3 (8 mg, tR = 10 min), which was identified as beauvericin (3) by comparing its physicochemical and spectroscopic data with its published values17; HR-ESI-MS: m/z 784.4163 [M+H]+ (calcd. C45H58N3O9, 784.4168); 1H-NMR (CD3OD, 400 MHz): δ 7.26 (4H, m, aromatic ring protons), 7.18 (1H, m, aromatic ring proton), 5.81 (1H, dd, J = 12.8, 4.4 Hz, CH-N), 4.84 (1H, d, J = 8.8 Hz, CH-O), 3.40 (1H, dd, J = 14.8, 4.4 Hz, CH2), 3.15 (3H, s, N-CH3), 3.04 (1H, dd, J = 14.8, 12.8 Hz, CH2), 1.80 (1H, m, CH), 0.86 (3H, d, J = 6.4 Hz, CH3), 0.24 (3H, d, J = 6.4 Hz, CH3); 13C-NMR (CD3OD, 100 MHz): δ 173.1 (O=C-N), 170.9 (O=C-O), 138.0 (aromatic ring), 129.8 (aromatic ring), 129.7 (aromatic ring), 128.0 (aromatic ring), 77.2 (CH-O), 57.8 (CH-N), 35.4 (CH2), 32.2 (N-CH3), 31.2 (CH), 19.1 (CH3), 17.2 (CH3).
Cyclosporin C (4) – Small-scale fungal extract of DS11- 10 (Chlamydocillium curvulum, 20 mg) was chromatographed by semi-preparative HPLC [C18, isocratic MeCN-H2O (85:15), flow rate: 4 mL/min] to yield compound 4 (9 mg, tR = 11 min), which was elucidated as cyclosporin C (4) by comparing its physicochemical and spectroscopic data with that of published values18; HR-ESI-MS: m/z 1218.8434 [M+H]+ (calcd. C62H112N13O11 1218.8436).
Tryptophol (5) – Compound 5 was isolated from a largescale fungal extract of DS8-6 (Aureobasidium melanogenum). DS8-6-F (fraction F of DS8-6 large-scale extract) was further subjected to preparative HPLC (C18, gradient elution with 30 to 95% MeOH in H2O with 0.1% formic acid, over 15 min using a 10 mL/min flow rate) to afford five subfractions (DS8-6-F-1~5). Compound 5 (4mg, tR = 22 min) was finally purified by semi-preparative HPLC [C18, isocratic MeCN-H2O (23:77), flow rate: 4 mL/min], and confirmed as tryptophol (5) by comparing its physicochemical and spectroscopic data with that of published values19; HR-ESI-MS: m/z 162.0914 [M+H]+ (C10H12NO, calcd. 162.0913); 1H-NMR (CD3OD, 400 MHz): δ 7.51 (1H, d, J = 8.0 Hz, H-4), 7.30 (1H, d, J = 8.0 Hz, H-7), 7.05 (1H, t, J = 7.2 Hz, H-6), 7.04 (1H, s, H-2), 6.96 (1H, t, J = 7.2 Hz, H-5), 3.78 (2H, t, J = 7.2 Hz, H-11), 2.95 (2H, t, J = 7.2 Hz, H-10).
Brefeldin A (6) – Brefeldin A (6) was obtained from a large-scale fungal extract of DS8-2 (Penicillium janthinellum). Compound 6 (20mg, tR = 11 min) was purified from DS8-2-G (30mg) by semi-preparative HPLC [C18, isocratic MeCN-H2O (35:65), flow rate: 4 mL/min], and determined as brefeldin A (6) by comparing its physicochemical and spectroscopic data with that of published values20; HRESI-MS: m/z 281.1747 [M+H]+ (C16H25O4, calcd. 281.1747); 1H-NMR (CD3OD, 400 MHz): δ 7.46 (1H, dd, J = 15.6, 3.2 Hz, H-3), 5.83 (1H, dd, J = 15.6, 2.0 Hz, H-2), 5.76 (1H, ddd, J = 15.2, 10.4, 4.4 Hz, H-11), 5.28 (1H, dd, J = 15.2, 9.6 Hz, H-10), 4.80 (1H, m, H-15), 4.22 (1H, m, H-7), 4.04 (1H, m, H-4), 2.39 (1H, m, H-9), 2.13 (1H, ddd, J = 13.6, 8.8, 5.2 Hz, H-8), 2.01 (1H, m, H-12), 1.99 (1H, m, H-6), 1.70-1.90 (5H, m, H-5,6,12,13,14), 1.59 (1H, m, H-14), 1.45 (1H, m, H-8), 1.24 (3H, d, J = 6.0 Hz, H-16), 0.89 (1H, m, H-13); 13C-NMR (CD3OD, 100 MHz): δ 168.3 (C-1), 155.1 (C-3), 138.1 (C-10), 131.4 (C-11), 117.7 (C-2), 76.6 (C-4), 73.2 (C-7), 73.0 (C-15), 53.2 (C-5), 45.4 (C-9), 44.1 (C-8), 41.8 (C-6), 35.0 (C-14), 33.0 (C-12), 28.0 (C-13), 21.1 (C-16).
Alternariol (7) – Small-scale fungal extract of DS8-3 (Alternaria brassicae, 20 mg) was purified by semipreparative HPLC [C18, isocratic MeCN-H2O (35:65), flow rate: 4 mL/min] to obtain alternariol (7) (3 mg, tR = 19 min)21; HR-ESI-MS: m/z 259.0599 [M+H]+ (calcd. C14H11O5, 259.0601); 1H-NMR (DMSO-d6, 400 MHz): δ 11.77 (OH-7), 10.92 (OH-9), 10.34 (OH-3), 7.24 (1H, d, J = 2.0 Hz, H-10), 6.71 (1H, d, J = 2.4 Hz, H-2), 6.64 (1H, d, J = 2.4 Hz, H-4), 6.37 (1H, d, J = 2.0 Hz, H-8), 2.70 (3H, s, H-11).
Terrein (8) – Small-scale fungal extract of DS1-13 (Aspergillus lentulus, 10 mg) was purified by semi-preparative HPLC [C18, isocratic MeCN-H2O (10:90), flow rate: 4 mL/min] to obtain terrein (8) (5 mg, tR = 10 min)22; HRESI-MS: m/z 155.0703 [M+H]+ (calcd. C8H11O3, 155.0703); 1H-NMR (CD3OD, 400 MHz): δ 6.83 (1H, dd, J = 15.6, 6.8 Hz, H-7), 6.44 (1H, d, J = 16.0 Hz, H-6), 6.00 (1H, s, H-5), 4.67 (1H, d, J = 2.8 Hz, H-3), 4.07 (1H, d, J = 2.8 Hz, H-2) 1.94 (1H, dd, J = 6.8, 1.6 Hz , H-8).
Penicillide (9) – Penicillide (9) was isolated from a largescale fungal extract of DS3-9 (Eurotiales sp.). Compound 9 (5 mg, tR = 18 min) was obtained from DS3-9-H (30 mg) by semi-preparative HPLC [C18, isocratic MeCN-H2O (50:50), flow rate: 4 mL/min]23; HR-ESI-MS: m/z 373.1646 [M+H]+ (calcd. C21H25O6, 373.1646); 1H-NMR (CD3OD, 400 MHz): δ 7.64 (1H, d, J = 8.4 Hz, H-2), 6.98 (1H, d, J = 8.4 Hz, H-1), 6.76 (1H, br s, H-10), 6.40 (1H, br s, H-8), 5.07 (2H, m, H-1ʹ), 5.05 (1H, m, H-7), 3.90 (3H, s, 4- OCH3), 2.20 (3H, s, 9-CH3), 1.75 (1H, m, H-3ʹ), 1.62 (1H, ddd, J = 14.0, 9.2, 5.2 Hz, H-2ʹ), 1.43 (1H, ddd, J = 14.0, 9.2, 4.4 Hz, H-2ʹ), 0.96 (3H, d, J = 6.8 Hz, H-4ʹ), 0.94 (3H, d, J = 6.8 Hz, H-5ʹ).
Dichlorodiaportin (10) – Dichlorodiaportin (10) was obtained from a large-scale fungal extract of DS2-7 (Trichoderma sp.). DS2-7-H (20 mg) was subjected to semi-preparative HPLC [C18, isocratic MeCN-H2O (50:50), flow rate: 4 mL/min] to give metabolite 10 (8 mg, tR = 16 min)24; HR-ESI-MS: m/z 319.0135 [M+H]+ (calcd. C13H13Cl2O5, 319.0135); 1H-NMR (CD3OD, 400 MHz): δ 6.51(2H, br s, H-4 and H-5), 6.47 (1H, br s, H-7), 6.03 (1H, br s, H-11), 4.30 (1H, m, H-10), 3.86 (3H, s, 6- OCH3), 2.98 (1H, br d, J = 14.8 Hz, H-9), 2.74 (1H, dd, J = 14.8, 9.2 Hz, H-9); 13C-NMR (CD3OD, 100 MHz): δ 167.2 (C-6), 166.1 (C-1), 163.3 (C-8), 153.5 (C-3), 139.5 (C-4a), 106.4 (C-4), 100.9 (C-5) , 100.2 (C-7), 99.4 (C-8a), 75.8 (C-11), 72.8 (C-10), 54.9 (O-Me), 35.9 (C-9).
Trichodermamide A (11) – Trichodermamide A (11) was isolated from a large-scale fungal extract of DS5-1 (Trichoderma harzianum). Compound 11 (10mg, tR = 16 min) was purified from DS5-1-G (20 mg) by semi-preparative HPLC [C18, isocratic MeCN-H2O (25:75), flow rate: 4mL/min]25; HR-ESI-MS: m/z 433.1241 [M+H]+ (calcd. C20H21N2O9, 433.1242); 1H-NMR (CD3OD, 400 MHz): δ 8.57 (1H, s, H-3ʹ), 7.29 (1H, d, J = 8.8, H-5ʹ), 7.06 (1H, d, J = 8.8 Hz, H-6ʹ), 5.60 (1H, d, J = 10.8 Hz, H-6), 5.53 (1H, d, J = 10.8 Hz, H-7), 4.38 (1H, br s, H-5), 4.14 (1H, m, H-8), 4.10 (1H, m, H-9), 3.92 (3H, s, 10ʹ-OCH3), 3.90 (3H, s, 11ʹ-OCH3), 2.67 (1H, d, J = 19.6 Hz, H-3), 2.24 (1H, d, J = 19.6 Hz, H-3); 13C-NMR (CD3OD, 100 MHz): δ 161.4 (C-1), 158.3 (C-1ʹ), 154.3 (C-7ʹ), 150.1 (C-2), 143.9 (C-9ʹ), 135.8 (C-8ʹ), 129.7 (C-6), 127.4 (C-7), 123.9 (C-3ʹ), 122.5 (C-5ʹ), 121.0 (C-2ʹ), 114.0 (C-4ʹ), 109.6 (C-6ʹ), 83.6 (C-9), 73.7 (C-5), 67.9 (C-4), 66.9 (C-8), 60.3 (C-11ʹ), 55.5 (C-10ʹ), 22.8 (C-3).
Radicinol (12) – Radicinol (12) was purified from a large-scale fungal extract of DS5-8 (Curvularia geniculata). DS5-8-F (50 mg) was subjected to semi-preparative HPLC [C18, isocratic MeCN-H2O (25:75), flow rate: 4 mL/min] to yield compound 12 (7 mg, tR = 12 min)26; HR-ESI-MS: m/z 239.0914 [M+H]+ (calcd. C12H15O5, 239.0914); 1H-NMR (CDCl3, 400 MHz): δ 6.69 (1H, dq, J = 15.6, 6.8 Hz, H-10), 5.94 (1H, dd, J = 15.6, 1.6 Hz, H-9), 5.74 (1H, s, H-8), 4.61 (1H, d, J = 7.2 Hz, H-4), 4.09 (1H, dq, J = 9.6, 6.4 Hz, H-2), 3.64 (1H, d, J = 9.6, 7.2 Hz, H-3), 1.88 (3H, dd, J = 6.8, 1.2 Hz, H-11), 1.49 (3H, d, J = 6.4 Hz, H-12); 13C-NMR (CDCl3, 100 MHz): δ 164.9 (C-5), 164.1 (C-8a), 158.7 (C-7), 135.6 (C-10), 122.5 (C-9), 100.3 (C-4a), 98.8 (C-8), 76.3 (C-2), 72.9 (C-3), 68.4 (C-4), 18.4 (C-11), 17.1 (C-12).
Sclerotioramine (13) – Sclerotioramine (13) was isolated from a large-scale fungal extract of DS2-11 (Penicillium sclerotiorum). DS2-11-H (50 mg) was subjected to semipreparative HPLC [C18, isocratic MeCN-H2O (70:30), flow rate: 4 mL/min] to obtain compound 13 (7 mg, tR = 7min)27; HR-ESI-MS: m/z 412.1280 [M+Na]+ (calcd. C21H24NNaO4, 412.1286); 1H-NMR (CDCl3, 400 MHz): δ 7.97 (1H, s, H-1), 7.08 (1H, d, J = 16.0 Hz, H-10), 6.91 (1H, s, H-4), 6.16 (1H, d, J = 16.0 Hz, H-9), 5.67 (1H, d, J = 10.0 Hz, H-12), 2.43 (1H, m, H-13), 2.12 (3H, s, H-20), 1.81 (3H, s, H-17), 1.53 (3H, s, H-18), 1.37 (1H, m, H-14), 1.27 (1H, m, H-14), 0.96 (3H, d, J = 6.8 Hz, H-16), 0.82 (3H, t, J = 7.2 Hz, H-15); 13C-NMR (CDCl3, 100MHz): δ 193.6 (C-8), 183.7 (C-6), 170.5, 148.5 (C-12), 147.5 (C-4a), 146.4 (C-3), 142.7 (C-10), 138.6 (C-1), 132.0 (C-11), 116.6 (C-9), 114.3 (C-8a), 110.7 (C-4), 101.1 (C-5), 85.0 (C-7), 35.0 (C-13), 30.0 (C-14), 23.5 (C-18), 20.3 (C-20), 20.2 (C-16), 12.4 (C-15), 11.9 (C-17).
Cell culture – MDA-MB-231 (triple-negative breast cancer) cell line was obtained from the Korean Cell Line Bank. MDA-MB-231 cells were cultured in RPMI-1640 (Corning Life Science) supplemented with 10% fetal bovine serum (FBS; Atlas Biologicals) and 1% penicillinstreptomycin (Gibco) and incubated in a humidified atmosphere containing 5% CO2 at 37oC.28
Cell Viability Assay – Cell viability was determined using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Cells were seeded in 96-well plates and incubated at 37oC for 24 h. Subsequently, cells were treated with various concentrations of the isolated compounds (0, 1, 5, 10, 20, and 40 μM) for 24 h. After treatment, 25 μL of 5 mg/mL MTT (Sigma-Aldrich, St. Louis, MO, USA) was added to each well and incubated for 3 h. The formazan crystals were dissolved using 100 μL of dimethyl sulfoxide. The optical density was measured at 540 nm using a Synergy 2 Multi-Mode Reader (BioTek Instruments Inc., Winooski, VT, USA).28
Results and Discussion
An in vitro cytotoxicity screening was performed using the MDA-MB-231 (triple-negative breast cancer) cells on the 96 fungal extracts (for a 96-well plate) isolated from 13 different soil samples, collected from the territory inside or near Duksung Women’s University such as gardens, lawns, and wetland in the university (Table 1). Among them, a subset of 22 extracts exhibited over 60% inhibition at 10 μg/mL compared to the negative control DMSO. Based on their HRESIMS data, the chemical profiling and dereplication were carried out, and five strains were excluded due to contamination and duplication. As a result, the top 17 hits were finally selected for purification of cytotoxic compounds (Table 3). For fungi producing multiple metabolites shown in the HR-ESI-MS data, active compounds were isolated through large-scale fermentation or will be studied later. Additionally, for several strains containing a single major metabolite, small-scale cultivation extracts for in vitro screening were subjected to one-step purification using HPLC semi-preparative methods.
Identification of the fungal strains corresponding to the 17 top-performing extracts revealed that five of them were identified as Trichoderma sp., which is a genus of soil-derived fungi well-known for producing peptaibols.29 Peptaibols are a class of linear peptidic fungal molecules possessing 5-20 amino acids, biosynthesized through the non-ribosomal peptide synthetase (NRPS) pathway.30 The sequences of these peptidic compounds (Pep-) mainly contain a-aminoisobutyric acid (-aib-), and a 2-amino alcohol (-ol) at the C-terminus.31 Peptaibols have been reported to exhibit a broad range of biological activities such as antimicrobial, antiviral, and anticancer, which are known to result from their amphipathic helical structures.32 This structural characteristic causes formation of ion channels in the bilayer membrane, ultimately leading to cell death.33 Large-scale cultivation of two strains producing peptaibols, DS7-1 (T. strigosum, 72.4% inhibition) and DS9-1 (T. strigosum, 71.1% inhibition), for chemical investigation led to the discovery of 13 and 12 new peptaibols, respectively (Fig. 3).15–16 These 27 compounds including 2 known molecules were tested for cytotoxicity against several cancer cell lines (MDA-MB-231, SKHep1, SNU449, SKOV3, DU145 and HCT116). Most of the isolated peptaibols showed potent inhibition activities against all tested cancer cell lines. Especially, among them, the most active compounds, strigaibol C (1) from DS7-1 and trichoguizaibol J (2) from DS9-1, exhibited sub-micromolar IC50 values of 0.99 μm and 0.68 μM against MDA-MB-231 cells, respectively (Table 4). Also, the other two strains DS9-4 (84.4% inhibition) and DS9-5 (82.9% inhibition) were both identified as T. spirale. Despite no peptaibols reported from this species yet, the presence of those molecules was detected by the dereplication process using HR-ESI-MS data. Thus, both fungi will be fermented in large-scale cultivation to find new cytotoxic compounds. The last remaining Trichoderma sp. strain out of the top-performing 17, DS6-6 (66.9% inhibition), was identified as T. harzianum. This fungus was recently researched by the Natural Products Discovery Group laboratory at the University of Oklahoma, where a total 27 peptaibols including 14 new ones were purified.34 A simple structure-activity relationship study based on their antiplasmodial activity revealed that one of 11 amino acid residue peptaibols (harzianin NPDG I) exhibited great activity (EC50 = 0.1 μM) and selectivity (> 250 compared with HepG2 EC50), and a possible mechanism of action was finally proposed.35 We also plan to obtain these peptaibols from DS6-6 in the near future and test their cytotoxicity against several cancer cell lines.
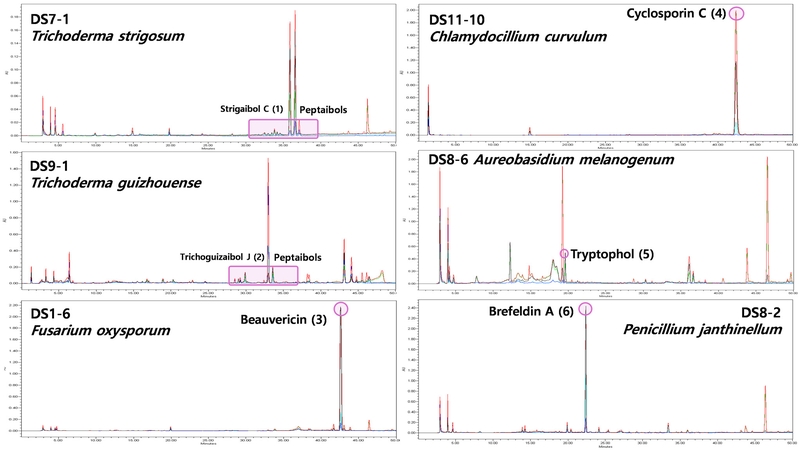
Initial HPLC analysis of top-performing fungal extracts (DS7-1, DS9-1, DS1-6, DS11-10, DS8-6 and DS8-2). Column: C18; Method: gradient elution 10%–100% MeCN in 40 min and isocratic 100% MeCN in 10 min.
Cell viability assay results against MDA-MB-231 cells (IC50 values expressed in μM ± SEM)a of compounds 1–13
DS10-9 strain identified as Epicoccum sorghinum exhibited great inhibitory activity of 81.7% against MDA-MB-231 cancer cells. To find the bioactive compounds thought as polyketides, large-scale cultivation was performed for isolation and purification.36 However, the major component, which was considered as the active one, gradually degraded over time, leading to failure in structure elucidation in the first attempt. We plan to carry out chemical investigation on the strain again through re-fermentation.
DS7-3 (71.9% inhibition, C. tenuissimum), DS9-3 (70.7% inhibition, C. perangustum), DS8-5 (62.9% inhibition, C. cladosporioides), and DS9-8 (60.9% inhibition, Cladosporium sp.), all identified as belonging to the genus Cladosporium have been reported to produce polyketides such as azaphilones and isocoumarins, and these strains were selected as candidates for a large-scale fermentation to find bioactive molecules.37–39 Another Cladosporium sp. strain (DS3-8) identified as C. anthropophilum showed 65% inhibitory activity. Its metabolites, however, did not seem to have been reported yet, so, this fungus will also be examined later.
DS1-6 fungus, identified as Fusarium sp., and DS11-10, identified as Chlamydocillium curvulum, were chemically dereplicated using HPLC, revealing the existence of a single outstanding major metabolite in the extracts of both strains (Fig. 3). For the evaluation of their cytotoxicity, these two major compounds were directly isolated from the small-scale fermentation extracts, and their structures were elucidated as beauvericin (3) and cyclosporin C (4) by interpreting the NMR and HRESIMS data, respectively.17,18 Since beauvericin (3), the major compound of the DS1-6 extract, showed a great IC50 value of 4.25 μM, it was presumed to be the active compound. In contrast, the active one of DS11-10, which exhibited 65.5% inhibition, was considered as minor components rather than cyclosporin C (4) with an IC50 of 96.3 μM.
The extracts of DS8-6 (Aureobasidium melanogenum) and DS8-2 (Penicillium janthinellum), isolated from slightly low-lying field soil next to the university library, showed 62.2% inhibition against MDA-MB-231 cells. Therefore, both fungi were cultivated on large scale, and we are currently isolating active compounds in those extracts. In the process, we first evaluated one of the isolated compounds, tryptophol (5) from DS8-6 and brefeldin A (6) from DS8- 2 (Fig. 3).19,20 As a result, tryptophol (5) exhibited mild activity with an IC50 of 83.6 μM, and we are continuing to find other more potent compounds and new biosurfactants in the extract of DS8-6. Brefeldin A (6) had an IC50 value of 38.2 μM, and we are also trying to search for new molecules with stronger activity from the extract of DS8-2.
DS3-7, isolated from soil in a forest established by the university decades ago, was identified as Penicillium simplicissimum. Chemical profiling using HPLC suggested DS3-7 mainly produces penicillic acid, which is a wellknown mycotoxin produced by Penicillium and Aspergillus sp.40 However, since various metabolites presumed as quinolone alkaloids were detected in the non-polar region (later than 25 min in our in-house HPLC method), we are currently in the process of large-scale cultivation and isolation of new molecules.41 DS6-2 is a strain isolated from a lawn near a dumpster, and was identified as Paraconiothyrium brasiliense, which is a genus known to produce sesquiterpenes such as brasilamides.42 As the extract of DS6-2 exhibited greater than 60% inhibitory effect against MDA-MB-231 cells, we are also preparing for large-scale cultivation to search for cytotoxic compounds.
Alternariol (7) and terrein (8) were purified from the small-scale extracts of DS8-3 and DS1-13, respectively (Fig. 4).21,22 While their extracts showed lower activity than the top-performing 17 extracts previously introduced, both compounds were evaluated for their cytotoxicity due to their inhibitory activity greater than 50% (Table 3). Based on the results, 7 showed no activity, whereas 8 exhibited mild activity with an IC50 value of 50.1 μM.
Initial HPLC analysis of underperforming fungal extracts (DS8-3, DS1-13, DS3-9, DS2-7, DS5-1, DS5-8 and DS2-11). Column: C18; Method: gradient elution 10%-100% MeCN in 40 min and isocratic 100% MeCN in 10 min.
Cell viability assay results against MDA-MB-231 cells (IC50 values expressed in μm± SEM, n = 3)a of compounds 1–13. aResults are expressed as means from triplicate experiments.; bPositive control.
Among the 96 extracts, 77 extracts that were not previously mentioned were dereplicated through HRESIMS analysis. As a result, 11 extracts were considered as candidates for large-scale cultivation, regardless of their MDA-MB-231 cell inhibition activities, to search for new molecules. Notably, four of these species (DS7-5, DS11- 2, DS3-6, and DS14-6) have not been researched for their metabolites yet (Fig. 4). Among 11 large-scale candidate fungi, five are currently undergoing large-scale cultivations for fractionation and isolation, and a major compound of each extract (9–13) was tested for cytotoxicity.23–27 The bioassay results revealed that compounds 9–11 had no or weak activities (IC50: >100 μM), while 12 and 13 exhibited mild activity (IC50: 54.2 μM) and strong activity with an IC50 value of 21.8 μM, respectively (Table 4). Based on the above-mentioned results, the analogues of 12 and 13, which are currently being obtained from DS5-8 and DS2-11 strains, respectively, are also expected to show cytotoxicity against MDA-MB-231 cells.
We isolated a total of 96 fungi from urban soil samples near Duksung Women’s University and carried out a cell viability screening using MDA-MB-231 cells to find cytotoxic compounds. Considering the diversity of natural product-derived compounds, the potential for redundancy in identities of strains and their metabolites, and statistical robustness, we simultaneously screened 96 extracts, a number that corresponds to commonly used well plates. Among the 96 small-scale-fermented fungal extracts, 17 strains that exhibited over 60% inhibitory activity at 10 μg/mL (vs DMSO) were selected as large-scale fermentation candidates to obtain cytotoxic metabolites. According to the results of a part of ongoing projects, six compounds (1–6) were isolated, and among them, the peptaibol series (1–2) and beauvericin (3) showed strong inhibition. Although the cytotoxicity in the screening results was lower than 60%, we also evaluated the inhibitory activity of major compounds (7–13), isolated from largescale cultivation fungal extracts selected for the discovery of new molecules through dereplication using HR-ESIMS data. As a result, compound 13 exhibited relatively great activity with an IC50 value of 21.8 μM.
The fungal metabolites isolated in this study have shown their potential as important sources of natural product-based anticancer drug development, and the strong cytotoxicity of certain compounds (peptaibol and beauvericin) supports their potential as novel drug candidates. Moreover, the relatively strong cytotoxicity of compound 13 suggests the potential for further enhancement through structural optimization and derivative development, indicating that it could be a promising lead compound for the development of novel anticancer drugs. Future research needs to conduct a more precise analysis of the mechanisms of action of these compounds and establish strategies for their integration into the novel drug development pipeline through an optimization process for preclinical research. Furthermore, we will continue our research to maximize the potential for developing natural product-based new drugs through the continuous isolation of bioactive compounds, structural identification of new molecules, and evaluation of cytotoxicity and other biological activities. We will also report on the cytotoxic activity screening of fungal extracts deposited in another 96-well plate.
Acknowledgments
This Research was supported by Duksung Women’s University Research Grants 2023-3000008345.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
REFERENCES
-
Abrahams, P. W. Sci. Total Environ. 2002, 291, 1–32.
[https://doi.org/10.1016/S0048-9697(01)01102-0]

-
Wurtzel, E. T.; Kutchan, T. M. Science 2016, 353, 1232–1236.
[https://doi.org/10.1126/science.aad2062]

-
Shara, M; Stohs, S. J. Phytother. Res. 2015, 29, 1112–1116.
[https://doi.org/10.1002/ptr.5377]

-
Weid, M.; Ziegler, J.; Kutchan, T. M. Proc. Natl. Acad. Sci. USA 2004, 101, 13957–13962.
[https://doi.org/10.1073/pnas.0405704101]

-
Wani, M. C.; Taylor, H. L.; Wall, M. E.; Coggon, P.; McPhail, A. T. J. Am. Chem. Soc. 1971, 93, 2325–2327.
[https://doi.org/10.1021/ja00738a045]

-
Raper, K. B.; Alexander, D. F.; Coghill, R. D. J. Bacteriol. 1944, 48, 639–659.
[https://doi.org/10.1128/jb.48.6.639-659.1944]

-
Waksman, S. A.; Reilly, H. C.; Johnstone, D. B. J. Bacteriol. 1946, 52, 393–397.
[https://doi.org/10.1128/jb.52.3.393-397.1946]

-
Omura, S.; Crump, A. Nat. Rev. Microbiol. 2004, 2, 984–989.
[https://doi.org/10.1038/nrmicro1048]

-
Jacoby, R.; Peukert, M.; Succurro, A.; Koprivova, A.; Kopriva, S. Front. Plant Sci. 2017, 8, 1617.
[https://doi.org/10.3389/fpls.2017.01617]

-
Li, L.; Yang, X.; Tong, B.; Wang, D.; Tian, X.; Liu, J.; Chen, J.; Xiao, X.; Wang, S. Front. Microbiol. 2023, 14, 1078886.
[https://doi.org/10.3389/fmicb.2023.1078886]

-
Du, L.; Robles, A. J.; King, J. B.; Powell, D. R.; Miller, A. N.; Mooberry, S. L.; Cichewicz, R. H. Angew. Chem. 2014, 126, 823–828.
[https://doi.org/10.1002/ange.201306549]

-
King, J. B.; Carter, A. C.; Dai, W.; Lee, J. W.; Kil, Y.-S.; Du, L.; Helff, S. K.; Cai, S.; Huddle, B. C.; Cichewicz, R. H. ACS Infect. Dis. 2019, 5, 1456–1470.
[https://doi.org/10.1021/acsinfecdis.9b00156]

-
Collins, J. E.; Lee, J. W.; Bohmer, M. J.; Welden, J. D.; Arshadi, A. K.; Du, L.; Cichewicz, R. H.; Chakrabarti, D. ACS Infect. Dis. 2021, 7, 2889–2903.
[https://doi.org/10.1021/acsinfecdis.1c00341]

-
Lee, J. W.; Collins, J. E.; Hulverson, M. A.; Aguila, L. K. T.; Kim, C. M.; Wendt. K. L.; Chakrabarti, D.; Ojo, K. K.; Wood, G. E.; Voorhis, W. C. V.; Cichewicz, R. H. J. Nat. Prod. 2023, 86, 1596–1605.
[https://doi.org/10.1021/acs.jnatprod.3c00283]

-
Park, Y. S.; Kim, E.-S.; Deyrup, S. T.; Lee, J. W.; Shim, S. H. J. Nat. Prod. 2024, 87, 2081–2094.
[https://doi.org/10.1021/acs.jnatprod.4c00590]

-
Han. J. S.; Kim, E.-S.; Cho, Y. B.; Kim. S. Y.; Lee, M. K.; Hwang. B. Y.; Lee, J. W. J. Nat. Prod. 2024, 87, 1994–2003.
[https://doi.org/10.1021/acs.jnatprod.4c00438]

-
Zhan, J.; Burns, A. M.; Liu, M. X.; Faeth, S. H.; Gunatilaka, A. A. L. J. Nat. Prod. 2007, 70, 227–232.
[https://doi.org/10.1021/np060394t]

-
Xu, L.; Li, Y.; Biggins, J. B.; Bowman B. R.; Verdine, G. L.; Gloer, J. B.; Alspaugh, J. A.; Bills, G. F. Appl. Microbiol. Biotechnol. 2018, 102, 2337–2350.
[https://doi.org/10.1007/s00253-018-8792-0]

-
Guzmán-López, O.; Trihos, Á.; Fernández, F. J.; Yañez-Morales, M. d. J.; Saucedo-Castañeda, G. World J. Microbiol. Biotechnol. 2007, 23, 1473–1477.
[https://doi.org/10.1007/s11274-007-9392-9]

-
Glaser, R.; Shiftan, D.; Froimowitz, M. Magn. Reson. Chem. 2000, 38, 274–280.
[https://doi.org/10.1002/(SICI)1097-458X(200004)38:4<274::AID-MRC630>3.0.CO;2-M]

-
Soukup, S. T.; Kohn, B. N.; Pfeiffer, E.; Geisen, R.; Metzler, M.; Bunzel, M.; Kulling, S. E. J. Agric. Food Chem. 2016, 64, 8892–8901.
[https://doi.org/10.1021/acs.jafc.6b03120]

-
Al Trabolsy, Z. B. K.; Anouar, E. H.; Zakaria, N. S. S.; Zulkeflee, M.; Hasan, M. H.; Zin, M. M.; Ahmad, R.; Sultan, S.; Weber, J.-F. J. Mol. Struct. 2014, 1060, 102–110.
[https://doi.org/10.1016/j.molstruc.2013.12.034]

-
Komai, S.-I.; Hosoe, T.; Itabashi, T.; Nozawa, K.; Yaguchi, T.; Fukushima, K.; Kawai K.-I. J. Nat. Med. 2006, 60, 185–190.
[https://doi.org/10.1007/s11418-005-0028-9]

-
Larsen, T. O.; Breinholt, J. J. Nat. Prod. 1999, 62, 1182–1184.
[https://doi.org/10.1021/np990066b]

-
Liu, R.; Gu, Q.-Q.; Zhu, W.-M.; Cui, C.-B.; Fan, G.-T. Arch. Pharm. Res. 2005, 28, 1042–1046.
[https://doi.org/10.1007/BF02977399]

-
Solfrizzo, M.; Vitti, C.; De Girolamo, A.; Visconti, A; Logrieco, A.; Fanizzi, F. P. J. Agric. Food Chem. 2004, 52, 3655–3660.
[https://doi.org/10.1021/jf035254t]

-
Wang, X.; Filho, J. G. S.; Hoover, A. R.; King, J. B.; Ellis, T. K.; Powell, D. R.; Cichewicz, R. H. J. Nat. Prod. 2010, 73, 942–948.
[https://doi.org/10.1021/np100142h]

-
Kim, E.-S.; Moon, A. Oncol. Lett. 2015, 9, 897–902.
[https://doi.org/10.3892/ol.2014.2735]

-
Marik, T.; Tyagi, C.; Balázs, D.; Urbán, P.; Szepesi, Á.; Bakacsy, L.; Endre, G.; Rakk, D.; Szekeres, A.; Andersson, M. A.; Salonen, H.; Druzhinina, I. S.; Vágvölgyi C.; Kredics. L. Front. Microbiol. 2019, 10, 1434.
[https://doi.org/10.3389/fmicb.2019.01434]

-
Leitgeb, B.; Szekeres, A.; Manczinger, L.; Vágvölgyi, C.; Kredics, L. Chem. Biodivers. 2007, 4, 1027–1051.
[https://doi.org/10.1002/cbdv.200790095]

-
Daniel, F. F. d. S.; Filho, E. R. Nat. Prod. Rep. 2007, 24, 1128–1141.
[https://doi.org/10.1039/b618086h]

-
Hou, X.; Sun, R.; Feng, Y.; Zhang, R.; Zhu, T.; Che, Q.; Zhang, G.; Li, D. Eng. Microbiol. 2022, 2, 100026.
[https://doi.org/10.1016/j.engmic.2022.100026]

-
Milov, A. D.; Tsvetkov, Y. D.; Raap, J.; Zotti, M. D.; Formaggio, F.; Toniolo, C. Pept. Sci. 2016, 106, 6–24.
[https://doi.org/10.1002/bip.22713]

-
Lee, J. W.; Collins, J. E.; Wendt, K. L.; Chakrabarti, D.; Cichewicz, R. H. J. Nat. Prod. 2021, 84, 503–517.
[https://doi.org/10.1021/acs.jnatprod.0c01370]

-
Collins, J. E.; Lee, J. W.; Rocamora, F.; Saggu, G. S.; Wendt, K. L.; Pasaje, C. F. A.; Smick, S.; Santos, N. M.; Paes, R.; Jiang, T.; Mittal, N.; Luth, M. R.; Chin, T.; Chang, H.; McLellan, J. L.; Morales-Hernandez, B.; Hanson, K. K.; Niles, J. C.; Desai, S. A.; Winzeler, E. A.; Cichewicz, R. H.; Chakrabarti, D. Cell Chem. Biol. 2024, 31, 312–325.e9.
[https://doi.org/10.1016/j.chembiol.2023.10.025]

-
Chang, C.-C.; Li, C.-Y.; Tsai, Y.-H.; El-Shazly, M.; Wei, C.-K.; Yang, Z.-J.; Chen, S.-L.; Wu, C.-C.; Wu. Y.-C.; Chang, F.-R. Planta 2021, 253, 116.
[https://doi.org/10.1007/s00425-021-03635-y]

-
Dai, H.-Q.; Kang, Q.-J.; Li, G.-H.; Shen, Y.-M. Helv. Chim. Acta 2006, 89, 527–531.
[https://doi.org/10.1002/hlca.200690055]

-
Fan, Z.; Sun, Z.-H.; Liu, H.-X.; Chen, Y.-C.; Li, H.-H.; Zhang, W.- M. J. Asian Nat. Prod. Res. 2016, 18, 1024–1029.
[https://doi.org/10.1080/10286020.2016.1181623]

-
Wang, X.; Radwan, M. M.; Tarawneh, A. H.; Gao, J.; Wedge, D. E.; Rosa, L. H.; Cutler, H. G.; Cutler, S. J. J. Agric. Food Chem. 2013, 61, 4551–4555.
[https://doi.org/10.1021/jf400212y]

-
Nguyen, H. T.; Yu, N. H.; Jeon, S. J.; Lee, H. W.; Bae, C.-H. Yeo, J. H.; Lee, H. B.; Kim, I.-S.; Park, H. W.; Kim, J.-C. Lett. Appl. Microbiol. 2016, 62, 488–493.
[https://doi.org/10.1111/lam.12578]

-
Dai, C.; Li, X.; Zhang, K.; Li, X.-N.; Wang, W.; Zang, Y.; Chen, X.; Li, Q.; Wei, M.; Chen, C.; Zhu, H.; Zhang, Y. Bioorg. Chem. 2021, 108, 104635.
[https://doi.org/10.1016/j.bioorg.2021.104635]

-
Liu, L.; Gao, H.; Chen, X.; Cai, X.; Yang, L.; Guo, L.; Yao, X.; Che, Y. Eur. J. Org. Chem. 2010, 17, 3302–3306.
[https://doi.org/10.1002/ejoc.201000284]