Cytotoxic Neoflavonoids and Chalcones from the Heartwood of Dalbergia melanoxylon
Abstract
Ten compounds, consisting of neoflavonoids (1-5), isoflavonoids (6 and 7), flavanone (8), and chalcones (9 and 10) were isolated from the ethyl acetate and n-butanol-soluble fractions of the heartwood of Dalbergia melanoxylon. The chemical structures were identified on the basis of spectroscopic evidence and compared to previously reported spectra. Compounds 1-10 were evaluated for cytotoxicity against HCT116 human colorectal cancer, MDA-MB-231 human metastatic breast cancer, and A2058 human melanoma cell lines. Among them, compounds 3 and 10 showed the strongest cytotoxic activity with IC50 values of 11.92 ± 1.07 μM, 10.83 ± 1.02 μM, and 14.37 ± 1.02 μM, 13.62 ± 1.09 μM against HCT116 and MDA-MB-231 cell lines, respectively. Compounds 9 and 10 also had cytotoxic activity with IC50 values of 13.49 ± 1.18 μM and 9.82 ± 0.91 μM against A2058 cell lines, respectively. To the best our knowledge, compounds 2 and 5-10 were isolated from this source for the first time.
Keywords:
Dalbergia melanoxylon, heartwood, HCT116, MDA-MB-231, A2058, neoflavonoid, chalconeIntroduction
During the screening for cytotoxic components from plants grown in Africa,1-4 Dalbergia melanoxylon (Guill. & Perr.) showed potent anti-proliferative activity on HCT116, MDA-MB-231 and A2058 cell lines. D. melanoxylon has been called by African people as “African Blackwood” or “Mpingo”. D. melanoxylon is high-profile species renowned for their use in high-quality products worldwide. D. melanoxylon is a small and heavily branched deciduous tree in the Family Leguminosae (= Fabaceae) that is widespread in at least 26 sub-Saharan countries including Tanzania, Kenya, Ethiopia and Nigeria.5,6 It normally reaches a height of 4-8 m, very occasionally 19-20 m, and often has multiple trunks. The wood has characteristically thin external yellowish white sapwood and internal purple heartwood. The hard, heavy wood is fine-grained, resistant to insect attack and is one of the most valuable timbers in Africa.7-9
Dalbergia species has various local medicinal uses (bark, roots and leaves) for the treatment of abdominal pain, joint pain, headache, hernia, gonorrhea, bronchitis,10 antimicrobial,6,11 cardioprotective,12 dermatitis,13 and brine shrimp toxicity.14 Various chemical constituents have been identified, including neoflavones,15,16 flavanones,17 benzofurans,18 N-cinnamoyl,19 quinone pigments,20 sesquiterpene,21 and phenanthrenedione22 from Dalbergia species.
However, the effects of components of D. melanoxylon on melanoma, breast and colorectal cancer activities have been rarely reported. Based on these backgrounds, the research was attempted to identify cytotoxic compounds from D. melanoxylon. This paper deals with the fractionation and isolation of ten constituents from the heartwood of D. melanoxylon, and the inhibitory effects of the them on the proliferation against HCT116 colorectal cancer, MDA-MB-231 breast cancer, and A2058 melanoma cell lines.
Experimental
General experimental procedures - The UV-visible spectra was recorded on a JP/U3010 Spectrophotometer (Hitachi Ltd., Tokyo, Japan). Nuclear magnetic resonance (NMR) spectra, including 1H-1H COSY, HMQC, and HMBC experiments were recorded on a Varian UI500 FT-NMR Spectrometer (Varian Inc, Palo Alto, CA, USA) operating at 500 MHz for 1H-NMR and 125 MHz for 13C-NMR with chemical shifts given in ppm (δ). Tetramethylsilane, acetone-d6, and DMSO-d6 were used as internal standard and NMR solvents. ESI-MS spectra was obtained on a JMS-AX 505 WA HP 5890 Series II, HP (Jeol, Tokyo, Japan). HPLC was carried out with JASCO model 887-PU pump an 875-UV variable-wavelength detector with a reversed-phase column (Develosil-Lop-ODS, 10-20 μm, 5 × 100 cm, Nomura Chemical co., Ltd, at 45 mL/min with detection at 205 nm). Column chromatography was run on a silica gel 60 (70-230 and 230-400 mesh, Merck, Darmstadt, Germany), C-18 (40-63 mesh, Merck), Diaion HP-20 (Merck) and Sephadex LH-20 (25-100 mm; GE Healthcare Life Sciences, Uppsala, Sweden). TLC was performed using Merck precoated silica gel 60F254 plates. Spots were detected on TLC under UV light and by heating after spraying with 10% H2SO4 in ethanol (v/v). NMR solvents were purchased from Sigma-Aldrich (St. Louis, MO, USA). 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and propidium iodide (PI) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), and 100 U/mL penicillin and 100 μg/mL streptomycin were purchased from Thermo Scientific Inc. (Waltham, MA, USA).
Plant Material - The heartwood of D. melanoxylon were collected at Zanzibar, East province, Tanzania in September 2013. The plant was identified by Professor Henry Joseph Hjndangalasi, Department of Botany, Dar es Salaam University, A voucher specimen (No. DH-201-10-47) was deposited in the herbarium of Dar es Salaam University, Dar es Salaam, Tanzania.
Extraction and Isolation - The dried heartwood of D. melanoxylon (600 g) was extracted with methanol (MeOH, 3 × 1.2 L) at room temperature for 3 days. The MeOH solution was evaporated to dryness to give 140.3 g of dark purple extracts. The MeOH extract was suspended in distilled water (600 mL) and partitioned with an equal volume of ethyl acetate (EtOAc), and then n-butanol (n-BuOH) successively, which afforded an EtOAc soluble fraction (32.6 g), n-BuOH soluble fraction (17.4 g) and remained aqueous fraction (20.1 g), respectively. The EtOAc soluble fraction (10 g) was subjected to silica gel column chromatographed (CC) (70-230 mesh), eluted with a gradient solvent system (MeOH in dichloromethane (CH2Cl2) 0% to 12%) to afford nine fractions (F1→F9). F1 (2.7 g) was subjected to silica gel CC (120 g, 230-400 mesh) and eluted with CH2Cl2-EtOAc (5% stepwise) to give seven subfractions (sub F1→sub F7). Sub F3 (320.8 mg) was chromatography on a silica gel eluted with n-hexane-EtOAc, and Sephadex LH-20 eluted with MeOH, and Diaion HP-20 eluted with MeOH-H2O to afford 1 (40.8 mg) and 5 (37.9 mg), while F2 (1.8 g) was chromatographed using silica gel CC (12 g, 230-400 mesh), eluted with a gradient solvent system (MeOH in CH2Cl2 5% to 35%) to afford five subfractions (sub F8→sub F12). Sub F9 (120.8 mg) was separated over RP-18 column chromatography with CHCl3: MeOH (10:1), which finally gave 8 (29.7 mg), and 9 (19.2 mg). Sub F11 (83.7 mg) was separated and crystallized with MeOH to yield 10 (22.8 mg).
F5 (2.4 g) was separated over silica gel CC (86 g, 70-230 mesh) with a gradient eluent of n-hexane-EtOAc-MeOH (gradient, 3:1:0→3:2:1) to produce seven subfractions (sub F13→19). Sub F14 (280.2 mg) and sub F17 (237.6 mg) were further separated over silica gel CC and eluted with EtOAc-MeOH (40:1) and EtOAc-MeOH (30:1), to afford sub F20→27 and sub F28→33, respectively. Subsequently, sub F23-26 (72.6 mg) was purified by Sephadex LH-20 eluted with MeOH to afford compound 2 (18.1 mg). Similarly, sub F29-30 (102.8 mg) and sub F31-32 (47.8 mg) were separated over silica gel CC (230-400 mesh), and eluted with n-hexane-EtOAc-MeOH (4:3:1) and EtOAc-MeOH (20:1) to afford 4 (18.1 mg) and 3 (12.7 mg), respectively. Sub F7 (142.3 mg) was separated over silica gel CC (230-400 mesh) and eluted with CH2Cl2-MeOH (82:18) to afford six sub fractions (sub F34→39). Sub F35-38 were purified with HPLC on Develosil-Lop. ODS (5 × 10 cm, 205 nm) eluted with acetone-H2O (45:55, 45 mL/min) to afford 6 (tR = 228 min, 16.1 mg), and eluted with acetone-H2O (20:80, 45 mL/min) to afford 7 (tR = 192 min, 12.5 mg), respectively.
Dalbergin (1) - White powder; ESI-MS, m/z 267.25 [M-H]-; 1H-NMR (acetone-d6, 500 MHz): δ 11.40 (1H, s, 6-OH), 7.59 (1H, m, H-4′), 7.48 (2H, m, H-3′,5′), 7.21 (2H, m, H-2′,6′), 7.08 (1H, s, H-8), 6.79 (1H, s, H-5), 6.16 (1H, s, H-3), 3.86 (3H, s, 7-OCH3); 13C-NMR (acetone-d6, 125 MHz): Table 1.
Dehydromelanoxin (2) - White powder; ESI-MS, m/z 299.30 [M-H]-; 1H-NMR (acetone-d6, 500 MHz): δ 7.86 (1H, s, 3′-OH), 7.34 (1H, s, 5-OH), 7.28 (1H, d, J = 2.4 Hz, H-2′), 7.21 (1H, dd, J = 2.4, 8.4 Hz, H-6′), 7.13 (1H, s, H-7), 7.04 (1H, d, J = 8.4 Hz, H-5′), 6.96 (1H, s, H-4), 3.91 (3H, s, 7-OCH3), 3.89 (3H, s, 4′-OCH3), 2.37 (3H, s, 3-CH3); 13C-NMR (acetone-d6, 125 MHz): Table 1.
Melanoxoin (3) - White powder; ESI-MS, m/z 289.27 [M-H]-; 1H-NMR (acetone-d6, 500 MHz): δ 13.08 (1H, s, 2-OH), 11.65 (1H br s, 3′-OH), 11.10 (1H, br s, 5-OH), 7.77 (1H, d, J = 2.0 Hz, H-2′), 7.69 (1H, s, H-6), 7.38 (1H, d, J = 2.0, 8.4 Hz, H-6′), 6.86 (1H, s, H-3), 6.75 (1H, d, J = 8.4 Hz, H-5′), 3.84 (3H, s, 4′-OCH3), 3.80 (3H, s, 4-OCH3); 13C-NMR (acetone-d6, 125 MHz): Table 1.
Melannein (4) - White powder; ESI-MS, m/z 313.30 [M-H]-, 1H-NMR (acetone-d6, 500 MHz): δ 11.61 (1H, br s, 3′-OH), 11.50 (1H, br s, 6-OH), 7.49 (1H, s, H-8), 7.31 (1H, dd, J = 2.1, 8.4 Hz, H-6′), 7.08 (1H, d, J = 8.4 Hz, H-5′), 7.07 (1H, s, H-5), 7.01 (1H, d, J = 2.1 Hz, H-2′), 6.44 (1H, s, H-3), 3.84 (3H, s, 4′-OCH3), 3.81 (3H, s, 7-OCH3); 13C-NMR (acetone-d6, 125 MHz): Table 1.
3′-Hydroxymelanettin (5) - White powder; ESI-MS, m/z 299.27 [M-H]-; 1H-NMR (acetone-d6, 500 MHz): δ 10.71 (1H, s, 3′-OH), 9.50 (1H, s, 4′-OH), 9.47 (1H, s, 6-OH), 7.38 (1H, dd, J = 2.4, 8.4 Hz, H-6′), 7.31 (1H, s, H-8), 7.12 (1H, s, H-5), 7.06 (1H, d, J = 8.4 Hz, H-5′), 6.98 (1H, d, J = 2.4 Hz, H-2′), 6.45 (1H, s, H-3), 3.81 (3H, s, 7-OCH3); 13C-NMR (acetone-d6, 125 MHz): Table 1.
Tectorigenin (6) - Pale yellow powder; ESI-MS, m/z 299.26 [M-H]-; 1H-NMR (acetone-d6, 500 MHz): δ 13.04 (1H, s, 5-OH), 10.74 (1H, s, 4′-OH), 9.58 (1H, s, 7-OH), 8.31 (1H, s, H-2), 7.28 (2H, d, J = 8.5 Hz, H-2′, H-6′), 6.81 (2H, d, J = 8.5 Hz, H-3′, 5′), 6.54 (1H, s, H-8), 3.71 (3H, s, 6-OCH3); 13C-NMR (acetone-d6, 125 MHz): Table 1.
Tectoridin (7) - Yellow plates; ESI-MS, m/z 461.43 [M-H]-; 1H-NMR (acetone-d6, 500 MHz): δ 13.04 (1H, s, 5-OH), 10.74 (1H, s, 4′-OH), 8.31 (1H, s, H-2), 7.35 (2H, d, J = 8.5 Hz, H-2′, H-6′), 6.83 (2H, d, J = 8.5 Hz, H-3′, 5′), 6.51 (1H, s, H-8), 4.65 (1H, d, J = 7.5 Hz, H-1″), 3.75 (3H, s, 6-OCH3), 3.69 (1H, m, H-6″b), 3.46 (1H, m, H-6″a), 3.40 (1H, m, H-3′′), 3.24 (1H, m, H-5″), 3.19 (1H, m, H-2″), 3.15 (1H, m, H-4″); 13C-NMR (acetone-d6, 125 MHz): Table 1.
Liquiritigenin (8) - Pale yellow plates; ESI-MS, m/z 255.23 [M-H]-; 1H-NMR (acetone-d6, 500 MHz): δ 7.67 (1H, d, J = 8.7 Hz, H-5), 7.53 (2H, d, J = 7.4 Hz, H-2′, 6′), 6.74 (1H, dd, J = 2.1, 6.4 Hz, H-6), 6.31 (1H, s, H-8), 6.08 (2H, d, J = 7.4 Hz, H-3′, 5′), 5.51 (1H, dd, J = 2.8, 12.8 Hz, H-2), 3.26 (1H, dd, J = 2.8, 12.8 Hz, H-3a), 2.65 (1H, dd, J = 2.8, 12.8 Hz, H-3b); 13C-NMR (acetone-d6, 125 MHz): Table 1.
2′-O-Methylisoliquiritigenin (9) - Colorless powder; ESI-MS, m/z 269.28 [M-H]-; 1H-NMR (DMSO-d6, 500 MHz): δ 10.1 (1H, br s, 4-OH), 9.86 (1H, br s, 4′-OH), 7.76 (1H, d, J = 15.0 Hz, H-β), 7.73 (1H, d, J = 15.2 Hz, H-α), 7.64 (1H, d, J = 8.6 Hz, H-6′), 7.51 (2H, d, J = 8.4 Hz, H-2, H-6), 7.40 (1H, d, J = 2.1 Hz, H-3′), 6.89 (2H, d, J = 8.4 Hz, H-3, H-5), 6.30 (1H, dd, J = 2.0, 8.4 Hz, H-5′), 3.85 (3H, s, 2′-OCH3); 13C-NMR (DMSO-d6, 125 MHz): Table 1.
Isoliquiritigenin (10) - White needles; ESI-MS, m/z 255.25 [M-H]-; 1H-NMR (acetone-d6, 500 MHz): δ 11.28 (3H, s, 2′-OH), 10.9 (1H, br s, 4-OH), 9.72 (1H, br s, 4′-OH), 7.73 (1H, d, J = 15.2 Hz, H-β), 7.64 (1H, d, J = 15.2 Hz, H-α), 7.60 (1H, d, J = 8.0 Hz, H-6′), 7.53 (2H, d, J = 8.4 Hz, H-2, H-6), 7.42 (1H, d, J = 2.0 Hz, H-3′), 6.89 (2H, d, J = 8.4 Hz, H-3, H-5), 6.32 (1H, dd, J = 2.0, 8.0 Hz, H-5′); 13C-NMR (acetone-d6, 125 MHz): Table 1.
Cell cultures - HCT116, MDA-MB-231, and A2058 cell lines were obtained from the Korean Cell Line Bank (KCLB, Seoul, Korea). The cells were maintained in Roswell Park Memorial Institute Media 1640 (RPMI 1640), supplemented with 10% FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin, and the cells were incubated at 37oC in a humidified incubator in a 5% CO2 atmosphere. Cell counts were performed using a hemocytometer from Hausser Scientific (Horsham, PA, USA).
Cytotoxicity assay - The cytotoxic effect of each compound against the HCT116, MDA-MB-231 and A2058 cell lines were estimated colorimetrical using MTT assay, which is based on the reduction of a tetrazolium salt by a mitochondrial dehydrogenase in viable cells.23 Doxorubicin (Sigma-Aldrich Co., >98.0%) was used as positive control. The results were expressed as relative viable cells compared to the controls (untreated cells). The 50% inhibitory concentration (IC50) values of anti-proliferative activity were calculated and converted into the IC50 Dose.
Results and Discussion
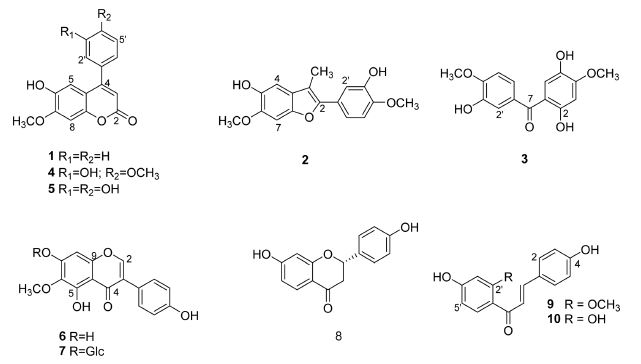
The MeOH extract of the dried heartwood of D. melanoxylon was partitioned successfully with EtOAc, n-BuOH and aqueous soluble fractions. Column chromatography was repeated using silica gel, Sephadex LH-20, and Diaion HP-20 of the soluble fractions of EtOAc and n-BuOH led to the isolation of ten compounds (1-10) (Fig. 1). The structures of the isolated compounds were identified by spectroscopic analysis including 1H,13C-NMR, and ESI-MS spectral data with those in the literatures to be dalbergin (1),24,25 dehydromelanoxoin (2),(25), melanoxoin (3),25 melannein (4),25 3′-hydroxymelanettin (5),24 tectorigenin (6),26-28 tectoridin (7),29 liquilitigenin (8),30,31 2′-O-methylisoliquilitigenin (9),31 and isoliquilitigenin (10)32,33 (Table 1). The isolated compounds 1-10 were evaluated for the cytotoxic activity against HCT116, MDA-MB-231, and A2058 cell lines through MTT assay, using Doxorubicin as a positive control. Among the compounds tested, compounds 2, 3, and 7-10 showed strong cytotoxicity with an IC50 values of < 30 μM against tested cell lines (Table 2).

Inhibition of tumor cell proliferation by compounds 1-10 against HCT116, MDA-MB-231, and A2058 cell lines
The naturally occurring neoflavonoids have been grouped together in accordance with their structural types and sources. Neoflavonoids are found in Leguminosae, Guttiferae, Rubiaeeae and more recently Passifloraceae. Among them, the arylcoumarins are of widespread distribution. The open-chain neoflavonoids can be subdivided according to their oxidation levels giving dalbergiquinol, dalbergione, and benzophenone groups. It is interesting to compare the different oxidation levels of isolated compounds from Dalbergia species.34 Up to date, arylcoumarin, arylbenzofuran, and benzophenone groups were isolated from the heartwood of Dalbergia melanoxylon. In the present study, compound 2 had a cytotoxicity with IC50 values of 21.64 ± 1.28 μM in A2058, and compound 3 of 11.92 ± 1.07 μM and 14.37 ± 1.02 μM in HCT116 and MDA-MB-231 cell lines, respectively. Compound 3, which has a carbonyl (C=O) linkage between the phenyl A and B rings exhibited cytotoxicity against HCT116 and MDA-MB-231 cell lines. Whereas, compounds 1, 4, and 5 which contain a lactone ring C and allyl (CH-CH=CH2) group formed by cyclization of a –C7-C8-C9-O- moiety onto ring B, was inactive (IC50 > 30.0 μM). These results were supported from previously studies that the presence of a carbonyl group in neoflavonoids structure can increase cytotoxic activity.25 Therefore, the structural comparison of compound 3 with the other neoflavonoids 1, 4, and 5 indicate that carbonyl linkage contribute cytotoxicity, whereas, heterocyclic ring C abolish to cytotoxicity in HCT116 and MDA-MB-231 cell lines.
Chalcone-based compounds (1,3-diaryl-2-propen-1-ones), natural or synthetic, have been widely reported to exhibit various biological activities, especially with regard to anticancer activities.35,36 Isoliquiritigenin (10) and its derivatives is applied into disease treatment such as cancer therapy and antibiotic therapy.37 In our experiments, compound 10 had a highest cytotoxicity against HCT116, MDA-MB-231, and A2058 cell lines of IC50 values of 10.83 ± 1.02 μM, 13.62 ± 1.09 μM, and 9.82 ± 0.91 μM, respectively. These results were supported from previously studies on cytotoxicity in colorectal cancer,32 and melanoma cell lines.38,39 Melanoma is the most aggressive type of skin cancer that originates from melanocytes, accounting for a high mortality rate annually.40 It is well known that surgical resection is the most common treatment for melanoma, but it is mostly invalid for patients with advanced melanoma, and the prognosis is poor with a 5-year survival rate < 16%.41 For these reasons, it has attracted increasing attention that looking for natural components and exploring the potential molecular mechanism of melanoma development is important for the treatment of malignant melanoma. Compound 9 also showed the anti-proliferative activity of IC50 values of 13.49 ± 1.18 μM in A2058 cell lines. On the other hand, compound 8 has rather inhibitory activity in HCT116 cell lines of IC50 values of 24.74 ± 1.62 μM, similar to published data.30 Colorectal cancer is thought to occur as a result of changes in the normal colon epithelial cells as adenomatous colorectal polyps. Chemotherapy and surgery are commonly used to treat cancer, but have many side effects. Flavonoids and related plant-derived phenolic compounds are well-known to have a wide range of cytotoxic activities. In the present study, compounds 3 and 10 might be intended to find natural resources with potent cytotoxic activity.
Overall, our research exhibited the heartwood of D. melanoxylon might be precious and valuable traded timber species. To the best our knowledge, compounds 2, and 5-10 were isolated from this source for the first time. However, further studies are required for investigation of cytotoxicity in molecular basis underlying in vitro and in vivo.
References
- Guon, T. E.; Yoon, K. M.; Chung, H. S. J. Kor. Tea Soc. 2015, 21, 59-64.
-
Guon, T. E.; Chung, H. S. Nat. Prod. Sci. 2016, 22, 209-215.
[https://doi.org/10.20307/nps.2016.22.3.209]

-
Guon, T. E.; Chung, H. S. Oncol. Lett. 2017, 14, 1703-1710.
[https://doi.org/10.3892/ol.2017.6288]

-
Guon, T. E.; Chung, H. S. Nat. Prod. Sci. 2017, 23, 227-234.
[https://doi.org/10.20307/nps.2017.23.4.227]

- Sacandé, M.; Vautier, H.; Sanon, M.; Schmit, L. SEED LEAFLET, No. 135; Dalbergia melanoxylon Guill. & Perr: UK, 2007.
-
Wang, M.; Ma, G.; Shao, F.; Liu, R.; Chen, L.; Liu, Y.; Yang, L.; Meng, X. Nat. Prod. Res. 2022, 36, 735-741.
[https://doi.org/10.1080/14786419.2020.1800692]

-
Li, Q.; Wu, J.; Wang, Y.; Lian, X.; Wu, F.; Zhou, L.; Huang, Z.; Zhu, S. Holzforschung 2017, 71, 939-949.
[https://doi.org/10.1515/hf-2017-0052]

- Jenkins, M.; Oldfield, S.; Aylett, T. International Trade in African Blackwood; Fauna and Flora International: UK, 2002, p 7.
-
Yin, X.; Huang, A.; Zhang, S.; Liu, R.; Ma, F. Molecules 2018, 23, 2163-2173.
[https://doi.org/10.3390/molecules23092163]

-
Mutai, P.; Heydenreich, M.; Thoithi, G.; Mugumbate, G.; Chibale, K.; Yenesew, A. Phytochem. Lett. 2013, 6, 671-675.
[https://doi.org/10.1016/j.phytol.2013.08.018]

-
Gundidza, M.; Gaza, N. J. Ethnopharmacol. 1993, 40, 127-130.
[https://doi.org/10.1016/0378-8741(93)90057-C]

-
Liu, Y.; Shu, J. C.; Wang, M. F.; Xu, Z. J.; Yang, L.; Meng, X. W.; Duan, W. B.; Zhang, N.; Shao, F.; Liu, R. H.; Chen, L. Y. Phytochem. 2021, 189, 112845.
[https://doi.org/10.1016/j.phytochem.2021.112845]

-
Vihervaara, A.; Liippo, J. Contact Dermatitis 2019, 81, 294-296.
[https://doi.org/10.1111/cod.13285]

-
Matata, D. Z.; Ngassapa, O. D.; Machumi, F.; Mainen, J. M. Evid. Based Complement. Alternat. Med. 2018, 2018, 3034612-3034627.
[https://doi.org/10.1155/2018/3034612]

-
Donnelly, D. M. X.; O’Reilly, J.; Whalley, W. B. Phytochem. 1975, 14, 2287-2290.
[https://doi.org/10.1016/S0031-9422(00)91118-X]

-
Gautam, J.; Kumar, P.; Kushwaha, P.; Khedgikar, V.; Choudhary, D.; Singh, D.; Maurya, R.; Trivedi, R. Menopause 2015, 22, 1246-1255.
[https://doi.org/10.1097/GME.0000000000000453]

-
Mutai, P.; Heydenreich, M.; Thoithi, G.; Mugumbate, G.; Chibale, K.; Yenesew, A. Phytochem. Lett. 2013, 6, 671-675.
[https://doi.org/10.1016/j.phytol.2013.08.018]

-
Donnelly, B. J.; Donnelly, D. M. X.; O’Sullivan, A. M.; Prendergast, J. P. Tetrahedron 1969, 25, 4409-4414.
[https://doi.org/10.1016/S0040-4020(01)82981-1]

-
Van Heerden, F. R.; Vincent Brandt, E.; Roux, D. G. Phytochem. 1980, 19, 2125-2129.
[https://doi.org/10.1016/S0031-9422(00)82207-4]

-
Barragán-Huerta, B. E.; Peralta-Cruz, J.; González-Laredo, R. F.; Karchesy, J. Phytochem. 2004, 65, 925-928.
[https://doi.org/10.1016/j.phytochem.2003.11.011]

-
Wang, H.; Dong, W. H.; Zuo, W. J.; Liu, S.; Zhong, H. M.; Mei, W. L.; Dai, H. F. Fitoterapia 2014, 95, 16-21.
[https://doi.org/10.1016/j.fitote.2014.02.013]

-
Lin, S.; Liu, R. H.; Ma, G. Q.; Mei, D. Y.; Shao, F.; Chen, L. Y. Nat. Prod. Res. 2020, 34, 2794-2801.
[https://doi.org/10.1080/14786419.2019.1591397]

- Carmichael, J.; DeGraff, W. G.; Gazdar, A. F.; Minna, J. D.; Mitchell, J. B. Cancer Res. 1987, 47, 936-942.
-
Chan, S. C.; Chang, Y. S.; Kuo, S. C. Phytochem. 1997, 46, 947-949.
[https://doi.org/10.1016/S0031-9422(97)00365-8]

-
Wu, S. F.; Chang, F. R.; Wang, S. Y.; Hwang, T. L.; Lee, C. L.; Chen, S. L.; Wu, C. C.; Wu, Y. C. J. Nat. Prod. 2011, 74, 989-996.
[https://doi.org/10.1021/np100871g]

-
Monthakantirat, O.; De-Eknamkul, W.; Umehara, K.; Yoshinaga, Y.; Miyase, T.; Warashina, T.; Noguchi, H. J. Nat. Prod. 2005, 68, 361-364.
[https://doi.org/10.1021/np040175c]

-
Umehara, K.; Nemoto, K.; Matsushita, A.; Terada, E.; Monthakantirat, O.; De-Eknamkul, W.; Miyase, T.; Warashina, T.; Degawa, M.; Noguchi, H. J. Nat. Prod. 2009, 72, 2163-2168.
[https://doi.org/10.1021/np900676y]

- Zeng, L.; Yuan, S.; Shen, J.; Wu, M.; Pan, L.; Kong, X. Mol. Med. Rep, 2018, 17, 3935-3943.
-
Xiong, L.; Guo, W.; Yang, Y.; Gao, D.; Wang, J.; Qu, Y.; Zhang, Y. Mol. Cell. Biochem. 2021, 476, 2729-2738.
[https://doi.org/10.1007/s11010-021-04081-w]

- Choi, C. W.; Choi, Y. H.; Cha, M. R.; Kim, Y. S.; Yon, G. H.; Kim, Y. K.; Choi, S. U.; Kim, Y. H.; Ryu, S. Y. J. Korean Soc. Appl. Biol. Chem. 2009, 52, 375-379.
-
Yahara, S.; Ogata, T.; Saijo, R.; Konishi, R.; Yamahara, J.; Miyahara, K.; Nohara, T. Chem. Pharm. Bull. 1989, 37, 979-987.
[https://doi.org/10.1248/cpb.37.979]

-
An, R. B.; Jeong, G. S.; Kim, Y.C. Chem. Pharm. Bull. 2008, 56, 1722-1724.
[https://doi.org/10.1248/cpb.56.1722]

- Guo, L. B.; Wang, L. Chin. Tradit. Herb. Drugs 2008, 39, 1147-1149.
-
Donnelly, D. M. X.; Sheriden, M. H. In The Flavonoid: Neoflavonoids; Harborne, J. B. Ed; Chapman & Hall, UK, 1988, pp 211-232.
[https://doi.org/10.1007/978-1-4899-2913-6_6]

-
Wattenberg, L. W.; Coccia, J. B.; Galbraith, A. R. Cancer Lett. 1994, 83, 165-169.
[https://doi.org/10.1016/0304-3835(94)90314-X]

-
Ducki, S.; Forrest, R.; Hadfield, J. A.; Kendall, A.; Lawrence, N. J.; McGown, A. T.; Rennison, D. Bioorg. Med. Chem. Lett. 1998, 8, 1051-1056.
[https://doi.org/10.1016/S0960-894X(98)00162-0]

-
Peng, F.; Du, Q.; Peng, C.; Wang, N.; Tang, H.; Xie, X.; Shen, J.; Chen, J. Phytother. Res. 2015, 29, 969-977.
[https://doi.org/10.1002/ptr.5348]

- Park, J. D.; Lee, Y. H.; Back, N. I.; Kim, S. I.; Ahn, B. Z. Kor. J. Pharmacogn. 1995, 26, 323-326.
-
Xiang, S.; Chen, H.; Luo, X.; An, B.; Wu, W.; Cao, S.; Ruan, S.; Wang, Z.; Weng, L.; Zhu, H.; Liu, Q. J. Exp. Clin. Cancer Res. 2018, 37, 184-199.
[https://doi.org/10.1186/s13046-018-0844-x]

-
Erdei, E.; Torres, S. M. Expert Rev. Anticancer Ther. 2010, 10, 1811-1823.
[https://doi.org/10.1586/era.10.170]

-
Haass, N. K.; Schumacher, U. Exp. Dermatol. 2014, 23, 471-472.
[https://doi.org/10.1111/exd.12400]