
Evaluation of Anti-cancer and Anti-proliferative Activity of Medicinal Plant Extracts (Saffron, Green Tea, Clove, Fenugreek) on Toll Like Receptors Pathway
Abstract
Despite considerable efforts, cancer remains an aggressive killer worldwide. Chemotherapeutic drugs that are currently in use lead to destructive side effects and have not succeeded in fulfilling expectations. For centuries, medicinal plants are used for treating various diseases and are also known to have anticancer activity. The main aim of this research was to evaluate antiproliferative activity of saffron, clove, fenugreek, and green tea on Vero and MDA-MB-231 cell lines and to subsequently analyze the effect of these extracts on IRAK-4, TAK1, IKK-alpha, IKK-beta, NF-Kappa B, IRF3, IRF7 genes in Toll Like Receptors (TLRs) pathway. Antiproliferative assay was done by Neutral Red Dye uptake assay. Methanolic extract of green tea was found to be most effective against both cell lines as IC50 was achieved at least concentration of the extract. For molecular studies, MDA-MB-231 cells were sensitized with methanolic extract of green tea at same IC50, and RT-PCR was performed to determine the relative expression of genes. Expression of IRAK-4, TAK1, IKK-beta, NF-Kappa B, IRF3 genes was down regulated and IRF7 and IKKalpha was upregulated. Green tea has a potential cytotoxic effect on both cell lines which was demonstrated by its effect on the expression of (TLRs) pathway genes.
Keywords:
Anti-proliferative effect, Cytotoxic effect, Toll Like Receptors (TLRs), NF-Kappa B, Medicinal plant extracts, green teaIntroduction
Many treatments are available to treat cancer, such as chemotherapy, radiotherapy, immunotherapy, targeted therapy, and hormone therapy.1 Despite growing research in this field and development of new drugs for cancer treatment, the efficacy of these drugs is challenged by cancerous cells. Cancer cells can develop resistance towards conventional drugs through mechanisms that promote or enable drug resistance, such as drug inactivation, drug target alteration, drug efflux, DNA damage repair, cell death inhibition, epithelial-mesenchymal transition and epigenetic modifications that can induce drug resistance. The increasing prevalence of these drug resistant cancer types necessitate further research and treatment development since available cancer treatments have several destructive side effects.2
Anti-metabolites and alkylating agents prevent cancer cell reproduction by causing direct damage to the DNA of cancer cells, however, this also causes long-term damage to the bone marrow which eventually leads to leukemia.3,4 Some chemotherapeutic agents cause peripheral neuropathy, brain, spinal cord and gait abnormalities and inability to concentrate.5 Free radicals generated by some chemotherapeutic agents’ cause DNA damage in the cells that frequently leads to apoptosis.6,7 Therefore, because of these pitfalls, there is a dire need of identifying treatment methods that are not harmful to host cells.
The biggest candidate for preventing and treating cancer are naturally occurring medicinal plants that have anticancer activity and are safe to use.8 Bioactive compounds present in the plants have shown promising results for the treatment and prevention of cancer in humans.9,10 Specifically speaking, various medicinal plants have been discovered which triggers pathways helpful in killing cancerous cells such as angiogenesis, JAK/STAT pathway and activation of apoptosis in cancer cells only.11 However, another potential pathway which can be targeted during cancer treatment is TLRs pathway. TLRs pathway plays an important role in maintaining tissue homeostasis by regulating the inflammatory and tissue repair responses to injury. Previously, it was reported that TLRs Pathway is involved in inducing inflammation which promotes carcinogenesis through promotion of angiogenesis and inhibition of apoptosis.12 However, studies observing the direct effect of plant extracts on TLRs for treatment of cancer are very limited. TLRs are a major target since they play a critical role in innate immune system. For this study, saffron, fenugreek, green tea, and clove extracts were used for the current study because in countries like Pakistan and India, aforementioned plants are found in abundance and are used in daily routine. Secondly, these plants have been traditionally used as anti-asthmatic, anti-inflammatory, anti-blood pressure, antioxidant, etc. More specifically, saffron and fenugreek only recently received attention for its numerous anticancer properties but not against TLRs. The aim of this study was hence two-folds. Firstly, effect of the chosen plants was tested on TLRs pathway to determine anticancer effect because only a handful of studies have observed the direct effect of these plants on TLRs and secondly, to identify the genes that are either up or downregulated by identifying the most effective plant extracts. For this, cytotoxic effect of the plant extracts was evaluated on Vero cancer cells lines and after confirming the effects on an animal cancer cell line, the results were consolidated by replicating on MDA-MB-231 human cancer cell line.
Experimental
Collection and Preparation of Plant Extracts – The plant samples were collected from local grocery stores (Al-Fatah and Hyperstar, Lahore, Pakistan) and voucher specimens were deposited in Botany Department of Kinnaird College for Women under voucher specimens (No.I412/2015-I3052/2017). The extracts of each plant (saffron, fenugreek, clove, and green tea) were prepared by using three different solvents (water, methanol, and ethanol). 5 g of each plant were homogenized per 5 mL of each type of solvent by using pestle and mortar. Extracts were then centrifuged for 15 minutes at 6000 rpm to remove the suspended particles. The supernatant of each sample was subjected to microfiltration by using 0 .2-micron filter under highly aseptic conditions in a laminar flow hood. The filtrate of each sample was collected in autoclaved Eppendorf and stored at -80ºC. Sterility of plant extract was tested by inoculating 100 μl of each extract in LB broth and the tubes were placed in the incubator for 24 hours. After 24 hours all tubes were clear and there was no growth in the broth which confirmed the sterility of the extracts.
Cell Cultures – Vero and MDA-MB-231 cell lines were obtained from Veterinary Research Institute (VRI), Lahore, Pakistan. Both cell lines were grown in Pyrex® Roux culture bottle in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS). The cells were maintained in humidifying atmosphere containing 5% CO2 at 37oC. All the cells were subcultured after 48-72 hours after evaluating the growth and morphology of cells under microscope.
Anti-proliferative Bioassays and IC50 Determination – To prepare 50 mL cell suspension, media of 3-4 flasks was discarded and monolayer was washed with phosphate buffered saline (PBS) twice. 10 mL PBS was added into each flask after which they were incubated at room temperature for 3-4 minutes. PBS was washed out and 1.5-2 mL of 2.5% Trypsin, 2% Versene solution was added and then the flask was incubated for 5-10 minutes until the cells were detached. The cells were examined under an Olympus IX81 inverted microscope to ensure that the cells remained detached and afloat. 6-8 mL of DMEM medium was added into each flask, after which the medium from all flasks were transferred into one flask. Lastly, the volume was made up to 50 mL by adding DMEM. After successful propagation, both the cell lines were used to study the inhibitory effect of the plant extracts prepared earlier. The antiproliferative bioassay was carried out in 96 wells flat bottom culture plates with lids.
Assay development – For the assay,100 μl of DMEM medium was added in all wells using an 8-channel micropipette. Then 100 μl extract was added in first well of the row and was 2-fold serially diluted by using 8-channel micropipette. After 2-fold dilution, 100 μl cell suspension was added to each well including last well. Last well was kept as a negative control consisting of DMEM medium and cell suspension. Ethanol, methanol, and water were also used on a separate 96 well plate, whereas the remaining protocol was performed as described above to rule out the sensitivity of the cell lines to these solvents which were run as control. The plates were placed in CO₂ incubator at 37oC for 48 hours and 5% CO₂ was provided for cell growth. The cell growth was monitored under microscope after 24 hours and upon completion of monolayer, the image of each well was captured by TIRF Imaging Microscope System.
Neutral Red Uptake Assay – The neutral red dye uptake assay was carried out to determine percentage viability of the cells in response to each plant extract. After 48 hours incubation, the media was removed from the 96-well plates and were washed with PBS thrice. After washing 100 μl (3.3 mg/mL) of neutral red dye was added to each well and incubated for 1 hour at 37oC. After incubation the dye was removed, and the wells were again washed with PBS thrice to remove any residual dye adhering to the outside of cells. The neutral red dye which was taken up by viable cells was extracted by adding 200 μl acidified ethanol (1 mL acetic acid, 50 mL ethanol, 49 mL distilled autoclaved water). The cells were kept in acidified ethanol for three minutes and absorbance was measured at 540 nm using a Microtiter plate reader to monitor cell growth in response to various concentrations of plant extract. This experiment was repeated thrice and the mean value ± SD was calculated. The absorbance against each of the fold dilution was recorded at 540 nm. The absorbance value of both the experimental and control was used to determine percentage of viable cells. This was done by using the equation below. Fold dilution of the plant sample that showed 50% viable cells were indicative of the IC50 value for that specific extract.
Total RNA Isolation – The effect of all plant extracts was observed on Vero and MDA-MB-231 cell lines. The concentration of extracts at which IC50 was achieved was calculated and that amount of extract was used to treat the cells. The cells were incubated at 37ºC for 48 hours. After incubation the monolayer of cells was split. The cell suspension was transferred to autoclaved Eppendorf and centrifuged at 12000 rpm for 5 minutes. The supernatant was discarded and 200 μl of RNA store reagent was added. The cell pellet was mixed well with the RNA store buffer. The Eppendorf were stored at -80ºC. The induced human cells were then subjected to RNA isolation.
To prepare the control/uninduced cells, the cells were grown without extract. The cells were incubated for 48 hours at 37ºC. After 48 hours, the cells were split and transferred into autoclaved Eppendorf and were then centrifuged at 12000 rpm for 5 minutes. The supernatant was discarded, and the pellet was then resuspended in the 200μl of RNA store buffer and were stored at -80ºC until further use.
RNA was isolated by using Thermo Scientific GeneJET RNA Purification Kit. RNA samples were treated with DNase prior to cDNA synthesis for the removal of any DNA contamination in RNA samples. To purify the RNA samples from DNA contamination, Tiangen DNA elimination Reagent Kit was used.
Quantitative Analysis of RNA – Determining the amount and purity of the RNA is very crucial for determining the amount of each sample to be used in downstream applications such as reverse transcription. Amplified products of treated and non-treated cells were evaluated to determine upregulation or downregulation. For this, quantitation of RNA was done by using nanodrop.
cDNA Synthesis – cDNA synthesis was carried out by using purified RNA as a template. RT-PCR was used to determine the relative expression of IRAK4, TAK1, IKK alpha, IKK beta, NF-kappa B, IRF3 and IRF7 genes along with the amplification of GAPDH which served as an internal control. RT-PCR and reaction mixture composition conditions were optimized to monitor expression profile of these genes in induced and uninduced MDA-MB-231 cells. Set of primers used for the amplification are shown in Table 1. The results of RT-PCR analysis were visualized on agarose gel.
Result and Discussion
Pattern of cell growth for both Vero and MDA-MB 231 cancer cell lines treated with decreasing concentration of plant extract were similar to the control and appeared to be elongated and polyhedral in shape. (Fig. 1). With increasing concentration of the extract, dead cells appeared which were rounded and oval with darker boundaries. The most effective plant extract in both cell lines was methanolic extract of green tea since cell viability was minimum at lowest concentration of plant extract (1024-fold dilution).
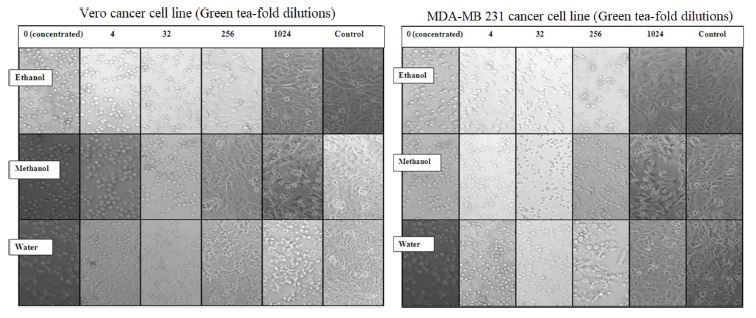
Vero cancer cell line and MDA-MB-231 cell proliferation pattern in response to green tea (Camellia sinensis) extracts. (Magnification 40X)
The absorbance of each well was recorded at 540 nm to monitor the cell growth in response to various concentrations of plant extracts. With reference to controls, results implied that with the increasing dilution of the extracts, there was gradual increase in the absorbance indicating cell viability. In Vero cell line, most effective plant extract was methanolic extract of green tea in since IC50 was achieved at least concentration of plant extract (1024-fold dilution). The least effective plant extract was ethanolic and water extracts of Saffron (16-fold dilution) (Table 3). According to the results, most effective plant extract in MDA-MB 231 cell line was methanolic extract of green tea since IC50 was achieved at least concentration of plant extract (1024-fold dilution). The least effective plant extract was water extract of Fenugreek (4-fold dilution) (Table 4).

Analysis of anti-proliferative effect of different dilutions of saffron, fenugreek, cove and green tea extracts on Vero cell line by neutral red uptake assay. IC50 is shown by percentages

Analysis of anti-proliferative effect of different dilutions of saffron, fenugreek, clove and green tea extracts on MDA-MB-231 cell line by neutral red uptake assay. IC50 is shown by percentages
IRF3 was observed at 323 bp and IRF7 at 515 bp. IRF3 was slightly downregulated in the treated cells and the expression level of IRF7 was upregulated in treated cells. IRAK4 was observed at 201 bp and expression level was observed to be downregulated in the treated cells. NFKappaB was observed at 285 bp and its expression level was downregulated in the treated cells. IKKalpha was observed at 290 bp and IKKbeta at 240 bp. It was observed that IKKalpha was slightly upregulated in treated cells while IKKbeta was downregulated in treated cells. TAK1 was observed at 198 bp and a slight downregulation was observed in the treated cells (Fig. 1 A-E).
In the current study, we studied the anti-proliferative activity of saffron, fenugreek, clove, and green tea on Vero and MDA-MB-231 cell lines to check the effect of these plants on cancer cells. The most effective plant extract was methanolic extract of green tea in both Vero and MDA-MB-231 cell lines which showed IC50 at least minimal dilution (1024). This result correlated with the work of Feng who investigated the anticancer and antioxidant effects of aqueous and ethanolic extracts of green tea that was enriched with selenium through MTT assay on HeLa cell line.13 Ethanolic extract of green tea inhibits the HeLa cell proliferation as compared to the aqueous extract and possess higher antitumor activity. It was observed that ethanolic extract as compared to aqueous extract was responsible for the greater antioxidant and antitumor activities. Second most effective plant extract was clove which can be considered as a potential anticancerous medical plant. Results revealed that methanolic extract and water extract of clove was most effective in case of Vero and MDA-MB 231 cell lines, respectively. This result correlated with the work of Dwivedi who observed the anticancer effects of clove in vitro by comparing three different extracts of clove on different cancer cell lines and found that cell growth was inhibited by the clove extracts and maximum cytotoxic activity was achieved by treating cells with oil extract of clove.14,15
We also found that the water and ethanolic of fenugreek was most effective when tested on Vero cells and MDA-MB-231 cells, respectively. Based on this, fenugreek can be considered as a potential anticancer agent as it showed potent cytotoxicity against cancer. In a similar study, the effect of fenugreek was studied on HT-29 human colon cancer cell line, and it was determined that fenugreek inhibits cell growth and induces apoptosis in the cells.16 In our study, least effective was methanolic extract of saffron against Vero cell lines while ethanolic extract was most effective against MDA-mb-231 cell line, as IC50 of these extracts was noted at 32-fold dilution. This finding correlated with the study of Bajbouj who reported that saffron induces the apoptosis and DNA damage in two p53 isogenic HCT116 cell lines which confirms its anticancer property.17
Results of gene expression (Fig. 2) revealed that green tea extract has a very strong effect on the genes of TLRs pathway. NFKappaB was downregulated in treated cells showing that inhibition of NFKappaB gene of TLRs pathway is critical as this gene plays an essential role in cancer development and progression and may also be involved in regulation of tumor angiogenesis and invasiveness.18 Since NFKappaB is the promoter of angiogenesis, its function can be inhibited by targeting its activator (IKKbeta) and inhibitor (IKKalpha) gene. In our study, IKKalpha gene (activator) was upregulated and IKKbeta gene (inhibitor) was downregulated in treated cells. As a result, IKKbeta gene activates IKKalpha gene which binds to the NFKappaB to inhibit its function. It was reported in different studies that IKKalpha and IKKbeta in addition to their roles in regulating immune responses through NFKappaB signaling cascades, also promotes tumor survival, proliferation, migration, metastasis, and angiogenesis-common characteristics of many types of human cancers. Hence, IKK and IKK-related kinases must be targeted for the development of novel therapeutic interventions for cancer.19
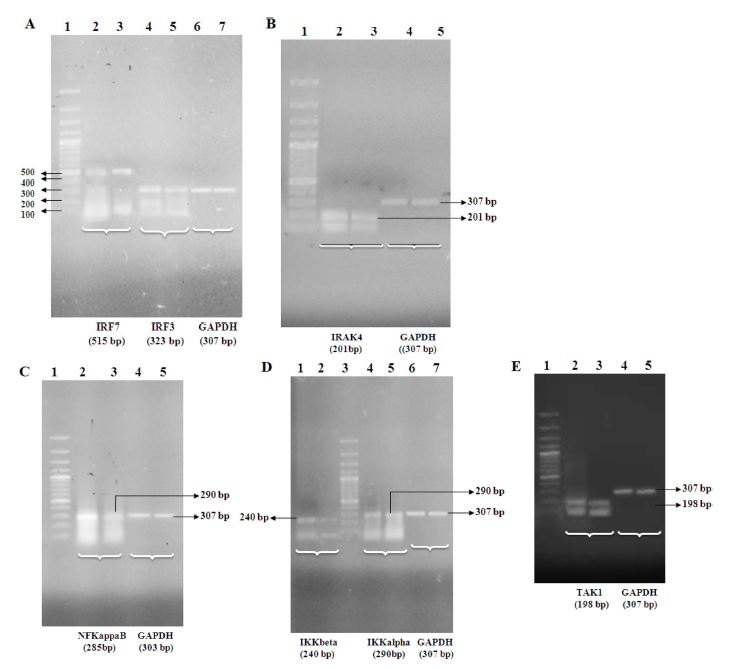
(A) Analysis of IRF3, IRF7 and internal control (GAPDH). Lane 1: Marker, Lane 2: Control IRF7, Lane3: Treated IRF7, Lane 4: Control IRF3, Lane 5: Treated IRF3, Lane 6: Control GAPDH, Lane 7: treated GAPDH (B) Analysis of IRAK4 gene and internal control (GAPDH). Lane 1: Marker, Lane 2: Control IRAK4, Lane 3: Treated IRAK4, Lane 4: Control GAPDH, Lane 5: Treated GAPDH. (C) Analysis of NFKappaB gene. Lane 1: Marker, Lane 2: NFKappaB Control, Lane3: Treated NFKappaB, Lane 4: Control GAPDH, Lane 5: Treated GAPDH. (D) Analysis of IKKalpha, IKKbeta genes and internal control (GAPDH). Lane 1: IKKbeta Control, Lane 2: Treated IKKbeta, Lane3: Marker, Lane 4: Control IKKalpha, Lane 5: Treated IKKalpha, Lane 6: Control GAPDH, Lane 7: Treated GAPDH. (E) Analysis of TAK1 gene and internal control (GAPDH). Lane 1: Marker, Lane 2: Control TAK1, Lane3: Treated TAK1, Lane 4: Control GAPDH, Lane 5: Treated GAPDH.
Furthermore, IRAK4 gene was downregulated in treated cells. IRAK4 plays a central role in pathways leading to cancer and inflammation by activating NF-κB. Therefore, IRAK4 can be a promising therapeutic target as inhibiting IRAK4 affect two processes i.e., inhibiting NFKappaB transcription and cancer cell survival and proliferation.20 TAK1 gene also showed the downregulation in treated cells. It was reported that TAK1 regulates nuclear factor-κB (NF-κB) that plays key roles in immune response and carcinogenesis. It was also demonstrated that TAK1 plays a role in tumor initiation, progression, and metastasis as a tumor prompter or tumor suppressor.21 Hence, targeting TAK1 will suppress tumor initiation. We also found that IRF3 was downregulated in treated cells and expression of IRF7 was slightly upregulated in treated cells. The IRF7 pathway was shown to be silenced in some metastatic breast cancer cell lines, which may help the cells avoid the host immune response.22 Restoring IRF7 in these cell lines reduced metastases and increased host survival time in animal models. Thus, tight regulation of IRF7 expression and activity is imperative in dictating appropriate type I IFN production for normal IFN-mediated physiological functions.23
In conclusion, this research through its beneficial outcomes provides key molecular targets (IRAK-4, TAK1, IKK-alpha, IKK-beta, NF-Kappa B, IRF3, and IRF7) for the development of most effective drug against cancer. As plants included in this study had significant cytotoxic and antiproliferative activity, these plant extracts can be clinically tested to produce the effective drugs for cancer treatment. The current study demonstrated that green tea is effective against the TLRs pathway genes and can be used as a therapeutic agent against cancer. Further studies are needed to isolate and study active component(s) in methanolic extract of green tea involved in the anti-proliferative effect. The effect of methanolic extract of green tea could be further tested in animal models and subsequently in human clinical trials.
References
-
Baskar, R.; Lee, K. A.; Yeo, R.; Yeoh, K. W. Int. J. Med. Sci. 2012, 9, 193-199.
[https://doi.org/10.7150/ijms.3635]

-
Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Cancers 2014, 6, 1769-1792.
[https://doi.org/10.3390/cancers6031769]

- Weber, G. F. Mol. Ther. Cancer 2014, 2014, 9-112.
-
Zajączkowska, R.; Kocot-Kępska, M.; Leppert, W.; Wrzosek, A.; Mika, J.; Wordliczek, J. Int. J. Mol. Sci. 2019, 20, 1451.
[https://doi.org/10.3390/ijms20061451]

-
Grisold, W.; Cavaletti, G.; Windebank, A. J. Neuro. Oncol. 2012, 14, iv45-iv54.
[https://doi.org/10.1093/neuonc/nos203]

-
Lobo, V.; Patil, A.; Phatak, A.; Chandra, N. Pharmacogn Rev. 2010, 4, 118-126.
[https://doi.org/10.4103/0973-7847.70902]

-
Dasari, S.; Tchounwou, B. P. Eur. J. Pharmacol. 2014, 740, 364-378.
[https://doi.org/10.1016/j.ejphar.2014.07.025]

- Malcolm R Alison. Encyclopedia of life sciences: Cancer; Nature Publishing Group: London, 2001, pp 1-8.
-
Patra, S.; Nayak, R.; Patro, S.; Pradhan, B.; Sahu, B.; Behera, C.; Bhutia, S. K.; Jena, M. Biotechnol. Rep (Amst). 2021, 30, e00633.
[https://doi.org/10.1016/j.btre.2021.e00633]

-
Kieliszek, M.; Edris, A.; Kot, A. M.; Piwowarek, K. Molecules 2020, 25, 2478.
[https://doi.org/10.3390/molecules25112478]

-
Mahmood, N.; Younas, H.; Zafar, M.; Shahid, S.; Ajmal, S.; Qursehi, Z.; Nasir, S. B. Nucleosides Nucleotides Nucleic Acids. 2021, 40, 434-469.
[https://doi.org/10.1080/15257770.2021.1896000]

-
O'Neill, L. A. J.; Golenbock, D.; Bowie, A. G. Nat. Rev. Immunol. 2013, 13, 453-460.
[https://doi.org/10.1038/nri3446]

-
Li, F.; Wang, F.; Yu, F.; Fang, Y.; Xin, Z.; Yang, F.; Xu, J.; Zhao, L.; Hu, Q. Food Chem. 2008, 111, 165-170.
[https://doi.org/10.1016/j.foodchem.2008.03.057]

- Dwivedi, V.; Shrivastava, R.; Hussain, S.; Ganguly, C.; Bharadwaj, M. Asian Pac. J. Cancer Prev. 2011, 12, 1989-1993.
- Abd El Azim, M. H. M.; EI-Mesallamy, A. M. D.; EI-Gerby, M.; Awad, A. J. Microb. Biochem. Technol. 2014, 10, s8-s007.
-
Khalil, M. I.; Tanvir, E. M.; Afroz, R.; Sulaiman, S. A.; Gan, S. H. Biomed. Res. Int. 2015, 2015, 286051.
[https://doi.org/10.1155/2015/286051]

-
Bajbouj, K.; Schulze-Luehrmann, J.; Diermeier, S.; Amin, A.; Schneider-Stock, R. BMC Complement. Altern. Med. 2012, 12, 69.
[https://doi.org/10.1186/1472-6882-12-69]

-
Karin, M. Cold Spring Harb Perspect Biol. 2009, 1, a000141.
[https://doi.org/10.1101/cshperspect.a000141]

-
Hoesel, B.; Schmid, J. A. Mol. Cancer 2013, 12, 86.
[https://doi.org/10.1186/1476-4598-12-86]

-
Rhyasen, G. W.; Starczynowski, D. T. Br. J. Cancer 2015, 112, 232-237.
[https://doi.org/10.1038/bjc.2014.513]

-
Roh, Y. S.; Song, J.; Seki, E. Br. J. Cancer 2014, 49, 185-194.
[https://doi.org/10.1007/s00535-013-0931-x]

-
Bidwell, B. N.; Slaney, C. Y.; Withana, N. P.; Forster, S.; Cao, Y.; Loi, S.; Andrews, D.; Mikeska, T.; Mangan, N. E.; Samarajiwa, S. A.; de Weerd, N. A.; Gould, J.; Argani, P.; Möller, A.; Smyth, M. J.; Anderson, R. L.; Hertzog, P. J.; Parker, B. S. Nat. Med. 2012, 18, 1224-1231.
[https://doi.org/10.1038/nm.2830]

-
Ning, S.; Pagano, J. S.; Barber, G. N. Genes Immun. 2011, 12, 399-414.
[https://doi.org/10.1038/gene.2011.21]
