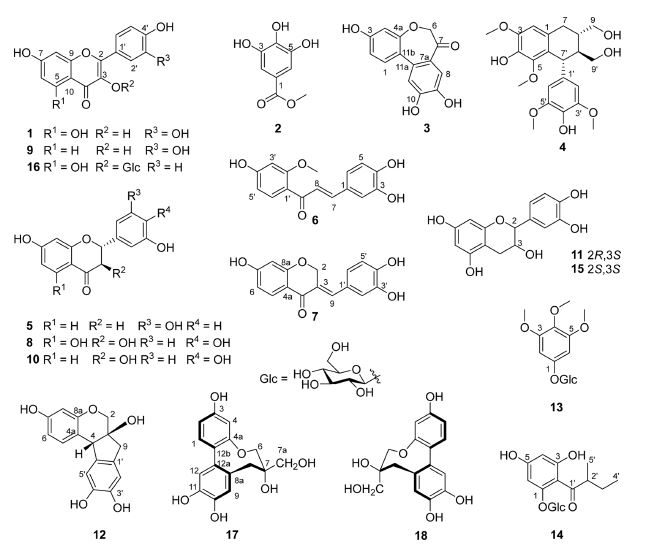
α-Glucosidase Inhibitory Activity of Phenolic Compounds Isolated from the Stems of Caesalpinia decapetala var. japonica
1College of Pharmacy, Drug Research and Development Center, Daegu Catholic University, Gyeongbuk 38430, Republic of Korea2College of Pharmacy, Research Institute of Pharmaceutical Science, Kyungpook National University, Daegu 41566, Republic of Korea
Abstract
In our study, sixteen known phenolic compounds, including quercetin (1), methyl gallate (2), caesalpiniaphenol C (3), 8S,8′S,7′R-(‒)-lyoniresinol (4), 7,3′,5′-trihydroxyflavanone (5), sappanchalcone (6), sappanone A (7), taxifolin (8), fisetin (9), fustin (10), (+)-catechin (11), brazilin (12), 3,4,5-trimethoxyphenyl β-ᴅ-glucopyranoside (13), 1-(2-methylbutyryl)phloroglucinol-glucopyranoside (14), (+)-epi-catechin (15), and astragalin (16) and one mixture of two conformers of protosappanin B (17/18) were isolated from the stems of Caesalpinia decapetala var. japonica. Their structures were elucidated based on a comparison of their physicochemical and spectral data with those of literature. To the best of our knowledge, this represents the first isolation of compounds 3, 4, 8, 9, and 10 from C. decapetala and compounds 13 and 14 from the Caesalpinia genus. All the isolated compounds were evaluated for their inhibitory effect against the α-glucosidase enzyme. Among them, two flavonols (1 and 9), one chalcone (6), and one homoisoflavanone (7) exhibited an inhibitory effect on α-glucosidase action with an IC50 range value of 5.08 ‒ 15.01 μM, stronger than that of the positive control (acarbose, IC50 = 152.22 μM). Kinetic analysis revealed that compounds 1 and 9 showed non-competitive α-glucosidase inhibition, while the inhibition type was mixed for compounds 6 and 7.
Keywords:
Caesalpinia decapetala var. japonica, Leguminosae, flavonoids, anti-diabetic, α-glucosidaseIntroduction
Diabetes, a chronic disease, is a heterogeneous metabolic disorder characterized by elevated blood glucose levels in the body. This disease affects millions of people around the world, causing a variety of life-threatening complications on cardiovascular, neurological, and kidney. Although conventional synthetic medicines are available, they are usually accompanied by serious adverse effects such as gastrointestinal problems disturbances, and metabolic effects.1 Thus, finding alternative natural medicine is an attractive strategy. The α -glucosidase enzyme hydrolyzes starch and sucrose down to glucose, inducing an increase in postprandial glucose levels in diabetic patients.2 Hence, inhibiting the activity of this enzyme produces a massive reduction of postprandial hyperglycemia, and decreases the risk of developing diabetes.3
The Caesalpinia genus (Leguminosae family) comprises approximately 100 species and spreads widely in the tropical and subtropical regions of Southeast Asia. Caesalpinia decapetala var. japonica is located in China, Korea, India, and Malaysia.4 Its whole plants had great medicinal values in folk medicine. Seeds of C. decapetala var. japonica were used for malaria and freckled skin.5 Flowers of this plant treat infusion in bronchitis, asthma, and malarial fevers.6 Leaves of C. decapetala var. japonica were used in the treatment of burns, biliousness, and stomach disorders.6 Its roots were applied to treat bronchitis, prevent colds, and anti-malarial.6 Roots and stems were also used for relieving pain.6 C. decapetala var. japonica contained various chemical components, including essential oils,7 flavonoids,8,9 chalcones,9 terpenoids,8,10 and steroids,9 which showed beneficial bioactivities such as anti-tumor,9 anti-oxidant,9 anti-analgesic,8 anti-inflammatory,8 anti-pyretic,8 and anti-virus.11
However, most of the previous phytochemical investigations of C. decapetala var. japonica focused on its root and leaves,12-14 the number of studies on its stems is still limited. Furthermore, although the blood-glucose-lowering effects of leaves and whole plant extract have been demonstrated,15,16 this effect of C. decapetala var. japonica stems and its chemical components have not been reported. Therefore, we herein conducted isolation and structural determination of chemical components from C. japonica stem extract, and investigation of their α-glucosidase inhibitory effect aimed to explore the anti-diabetic activity of C. decapetala var. japonica stems.
Experimental
General experiment procedures ‒ Specific optical rotation was measured using a JASCO P-2000 digital polarimeter (JASCO Corporation, Tokyo, Japan). The 1D and 2D-NMR spectra were obtained on a Varian Unity Inova 400 MHz (Varian, Inc., California, USA) spectrometer with tetramethylsilane (TMS) as an internal standard and the chemical shifts were recorded in δ values (ppm). Silica gel (Merck, 0.040-0.063 mm), RPC-18 (Merck, 17 mesh), and Sephadex LH-20 (Pharmacia Company) were used for column chromatography (CC). High-performance liquid chromatography was carried out on a Waters 2487 controller system with a UV detector (UV/VIS-156). RPC-18 F254s and silica gel F254 (Merck) plates were used to perform thin-layer chromatography (TLC). Compounds were detected under UV light and then visualized by heating after spraying with H2SO4 10%.
Chemical and reagents ‒ Yeast α-glucosidase, p-nitrophenyl α-ᴅ-glucopyranoside (p-NPG), sodium phosphate, sodium carbonate, acarbose, and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich Co. (St. Louis, MO, U.S.A).
Plant materials ‒The stems of Caesalpinia decapetala var. japonica were collected at Jeju-do, Republic of Korea, in June 2018 and identified by Professor Byung Sun Min. A voucher specimen (CUD-1857-1) was deposited at the Herbarium of the College of Pharmacy, Daegu Catholic University, Korea.
Extraction and isolation ‒ The dried stems of C. decapetala var. japonica (16.0 kg) were extracted with MeOH (20 L × 5 times × 4 h) and then evaporated to dryness to obtain the MeOH extract. The MeOH extract (4.0 kg) was suspended in distilled water (3 L) and then partitioned successively with n-hexane, CH2Cl2, EtOAc, and n-butanol to collect n-hexane (715.1 g), CH2Cl2 (408.5 g), EtOAc (110.1 g), and n-butanol (490.2 g) fractions and a water residue, respectively.
The EtOAc fraction (110.1 g) was chromatographed over a silica gel column (CH2Cl2:MeOH, stepwise, 25:1→0:1) to collect 4 fractions (E1‒E4). E2 (15.5 g) was then separated by a silica gel CC (CH2Cl2:MeOH, 20:1) to yield 7 sub-fractions E2.1‒E2.7. Fraction E2.4 (3.1 g) was fractionated by a normal phase CC (CH2Cl2:MeOH, 25:1) to furnish compound 1 (6.0 mg) and sub-fractions E2.4.1‒E3.4.5. E2.4.3 (234.2 mg) was purified by RPC-18 CC (MeOH:H2O, 1:3) and then Sephadex LH-20 CC (MeOH: H2O, 1:1) to afford 2 (5.1 mg) and 3 (9.8 mg). By the same method, compounds 4 (9.9 mg), 5 (30.1 mg), and 6 (49.5 mg) were collected from E2.4.4 (345.5 mg), and compounds 7 (6.0 mg) and 8 (50.1 mg) were obtained from E2.4.5 (158.7 mg). Fractionation of E2.6 passed over silica gel CC (CH2Cl2:MeOH, 22:1) to yield subfractions E2.6.1‒E2.6.5. Sub-fraction E2.6.4 (359.7 mg) was subjected to chromatography on a RPC-18 column (MeOH:H2O, 1:4) and subsequently purified by Sephadex LH-20 CC (MeOH:H2O, 1:1) to obtain 9 (15.2 mg) and 10 (80.6 mg). Sub-fraction E2.6.5 (390.1 mg) was separated by RPC-18 CC using MeOH: H2O (1:4) and further purified by a preparative Waters HPLC system (40% MeOH in 5 mL/min, 40 min) to yield 11 (3.0 mg, tR = 28.4 min). Compound 12 (5.1 mg) was afforded from E2.7 (124.6 mg) under the same conditions. E3 (4.5 g) was divided into 8 sub-fractions E3.1‒E3.8 by using a silica gel CC (CH2Cl2: MeOH, 12:1). Fraction E3.3 (556.7 mg) was continuously separated by a silica gel CC (CH2Cl2: MeOH, 10:1) to give 5 sub-fractions E3.3.1‒E3.3.5. Subfraction E3.3.5 (159.7 mg) was chromatographed over a RPC-18 CC (MeOH:H2O, 1:3) and then purified by Sephadex LH-20 CC (MeOH:H2O, 1:2) to furnish 13 (14.5 mg). Fraction E3.7 (631.4 mg) was divided into 3 sub-fractions E3.7.1‒E3.7.3 by a silica gel CC (CH2Cl2:MeOH, 9:1). Application the same method in case of subfraction E2.6.4 for sub-fraction E3.7.2 (324.2 mg), compounds 14 (80.0 mg) and 15 (9.7 mg) were isolated. Similarly, compound 16 (5.0 mg), and a mixture 17/18 (30.1 mg) were afforded from E3.8.3 (250.4 mg).
Quercetin (1) ‒ Amorphous yellow powder; C15H10O7; 1H NMR (400 MHz, CD3OD): δH 7.71 (1H, s, H-2′), 7.61 (1H, d, J = 8.4 Hz, H-6′), 6.86 (1H, d, J = 8.4 Hz, H-5′), 6.36 (1H, s, H-6), 6.16 (1H, s, H-8); 13C NMR (100 MHz, CD3OD): δC 175.9 (C-4), 164.2 (C-7), 161.0 (C-5), 156.8 (C-9), 147.3 (C-4′), 146.5 (C-3′), 144.8 (C-2), 135.8 (C-3), 122.7 (C-1′), 120.2 (C-6′), 114.8 (C-5′), 114.6 (C-2′), 103.0 (C-10), 97.8 (C-6), 92.9 (C-8).17
Methyl gallate (2) ‒ Amorphous white powder; C8H8O5; 1H NMR (400 MHz, CD3OD): δH 7.02 (2H, s, H-2, 6), 3.79 (3H, s, 7-OCH3); 13C NMR (100 MHz, CD3OD): δC 167.6 (C-7), 145.0 (C-3, 5), 138.3 (C-4), 119.9 (C-1), 108.6 (C-2, 6), 50.8 (7-OCH3).18
Caesalpiniaphenol C (3) ‒ Yellow needles; C14H10O5; 1H NMR (400 MHz, CD3OD): δH 7.06 (1H, d, J = 8.2 Hz, H-1), 6.68 (2H, s, H-8, 11), 6.66 (1H, dd, J = 8.4, 1.8 Hz, H-2), 6.62 (1H, d, J = 1.8, H-4), 4.42 (2H, s, H-6); 13C NMR (100 MHz, CD3OD): δC 207.7 (C-7), 159.6 (C-3), 159.0 (C-4a), 145.6 (C-9), 145.3 (C-10), 131.8 (C-11a), 130.9 (C-1), 127.1 (C-11b), 124.8 (C-7a), 117.4 (C-8, C-11), 113.3 (C-2), 108.8 (C-4), 78.8 (C-6).19
(8S,8′S,7′R)-(‒)-lyoniresinol (4) ‒ Amorphous white powder; C22H28O8; ‒40 (c = 0.025, MeOH); 1H NMR (400 MHz, CD3OD): δH 6.56 (1H, s, H-2), 6.36 (2H, s, H-2′, 6′), 4.29 (1H, d, J = 5.7 Hz, H-7′), 3.83 (3H, s, 3-OCH3), 3.71 (6H, s, 3′, 5′-OCH3), 3.64 (1H, d, J = 5.5 Hz, Hb-9′), 3.57 (1H, dd, J = 10.8, 5.0 Hz, Hb-9), 3.48 (2H, m, Ha-9′, Ha-9), 3.35 (3H, s, 5-OCH3), 2.68 (1H, dd, J = 15.0, 4.8 Hz, Hb-7), 2.55 (1H, dd, J = 15.0, 11.8 Hz, Ha-7), 1.95 (1H, dd, J = 8.1, 5.5 Hz, H-8′), 1.61 (1H, m, H-8); 13C NMR (100 MHz, CD3OD): δC 147.5 (C-3′, 5′), 147.2 (C-3), 146.2 (C-5), 137.8 (C-1′), 137.4 (C-4), 133.0 (C-4′), 128.7 (C-1), 124.8 (C-6), 106.3 (C-2), 105.4 (C-2′, 6′), 65.3 (C-9), 62.7 (C-9′), 58.7 (5-OCH3), 55.3 (3-OCH3), 55.2 (3′, 5′-OCH3), 47.6 (C-8′), 40.9 (C-7′), 39.4 (C-8), 32.1 (C-7).20
7,3′,5′-Trihydroxyflavanone (5) ‒ Amorphous white powder; C15H14O5; 1H NMR (400 MHz, CD3OD): δH 7.69 (1H, dd, J = 8.8, 0.8 Hz, H-5), 6.91 (1H, s, H-4′), 6.77 (2H, s, H-2′, 6′), 6.47 (1H, d, J = 8.8 Hz, H-6), 6.33 (1H, d, J = 1.2 Hz, H-8), 5.28 (1H, d, J = 12.8 Hz, H-2), 2.98 (1H, dd, J = 16.8, 12.8 Hz, Ha-3), 2.66 (1H, dd, J = 16.8, 2.4 Hz, Ha-3); 13C NMR (100 MHz, CD3OD): δC 192.2 (C-4), 165.2 (C-7), 164.1 (C-9), 145.3 (C-5′), 145.0 (C-3′), 130.1 (C-1′), 128.4 (C-5), 117.8 (C-2′), 114.8 (C-6′), 113.5 (C-10), 113.3 (C-4′), 110.3 (C-6), 102.4 (C-8), 79.6 (C-2), 43.5 (C-3).21
Sappanchalcone (6) ‒ Amorphous yellow powder; C16H14O5; 1H NMR (400 MHz, CD3OD): δH 7.55 (1H, d, J = 8.5 Hz, H-6′), 7.47 (1H, d, J = 15.6 Hz, H-7), 7.34 (1H, d, J = 15.6 Hz, H-8), 7.09 (1H, s, H-2), 6.96 (1H, d, J = 8.2 Hz, H-6), 6.77 (1H, d. J = 8.2 Hz, H-5), 6.48 (1H, d, J = 1.6 Hz, H-3′), 6.43 (1H, d, J = 8.5, 1.6 Hz, H-5′), 3.86 (3H, s, 2′-OCH3); 13C NMR (100 MHz, CD3OD): 191.7 (C-9), 163.1 (C-4′), 161.1 (C-2′), 148.1 (C-4), 145.4 (C-3), 143.1 (C-7), 132.3 (C-6′), 127.2 (C-1), 123.7 (C-8), 121.9 (C-6), 120.3 (C-1′), 115.1 (C-5), 113.8 (C-2), 107.5 (C-5′), 98.7 (C-3′), 54.7 (2′-OCH3).22
Sappanone A (7) ‒ Amorphous yellow powder; C16H12O5; 1H NMR (400 MHz, CD3OD): δH 7.77 (1H, d, J = 8.7 Hz, H-5), 7.62 (1H, s, H-9), 6.84 (1H, d, J = 8.1 Hz, H-5′), 6.80 (1H, s, H-2′), 6.74 (1H, d, J = 8.1 Hz, H-6′), 6.50 (1H, d, J = 8.7 Hz, H-6), 6.28 (1H, s, H-8), 5.33 (2H, s, H-2); 13C NMR (100 MHz, CD3OD): 181.6 (C-4), 165.1 (C-7), 163.4 (C-8a), 147.3 (C-3′), 145.1 (C-4′), 137.0 (C-9), 129.2 (C-5), 128.0 (C-1′), 126.2 (C-3), 123.1 (C-6′), 116.9 (C-5′), 115.1 (C-2′), 114.5 (C-4a), 110.7 (C-6), 102.2 (C-8), 67.7 (C-2).23
Taxifolin (8) ‒ Amorphous colorless powder; C15H12O7; 1H NMR (400 MHz, CD3OD): δH 6.95 (1H, d, J = 1.5 Hz, H-2), 6.83 (1H, dd, J = 8.1, 1.5 Hz, H-6), 6.78 (1H, d, J = 8.1 Hz, H-5′), 5.90 (1H, d, J = 1.5 Hz, H-8), 5.86 (1H, d, J = 1.5 Hz, H-6), 4.87 (1H, d, J = 11.5 Hz, H-2), 4.48 (1H, d, J = 11.5 Hz, H-3); 13C NMR (100 MHz, CD3OD): δC 196.9 (C-4), 167.3 (C-7), 163.8 (C-5), 163.0 (C-9), 145.7 (C-4′), 144.8 (C-3′), 128.4 (C-1′), 119.6 (C-6′), 114.7 (C-5′), 114.5 (C-2′), 100.4 (C-10), 95.9 (C-6), 94.9 (C-8), 83.6 (C-2), 72.2 (C-3).24
Fisetin (9) ‒ Amorphous white powder; C15H10O7; 1H NMR (400 MHz, CD3OD): δH 7.96 (1H, dd, J = 9.4, 0.5 Hz, H-5), 7.75 (1H, d, J = 2.1 Hz, H-2′), 7.65 (1H, ddd, J = 8.5, 2.1, 0.5 Hz, H-6′), 6.88 (3H, m, H-6, H-8, H-5′); 13C NMR (100 MHz, CD3OD): δC 172.9 (C-4), 162.9 (C-7), 157.1 (C-9), 147.2 (C-4′), 146.0 (C-3′), 144.8 (C-2), 137.1 (C-3), 126.0 (C-5), 122.9 (C-1′), 120.2 (C-6′), 114.8 (C-5′), 114.5 (C-6), 114.5 (C-2′), 113.9 (C-10), 101.5 (C-8).25
Fustin (10) ‒White needles; C15H12O6; 1H NMR (400 MHz, CD3OD): δH 7.69 (1H, d, J = 8.7 Hz, H-5), 6.97 (1H, d, J = 1.9 Hz, H-2′), 6.84 (1H, dd, J = 8.1, 1.9 Hz, H-6′), 6.79 (1H, d, J = 8.1, H-5′), 6.50 (1H, dd, J = 8.7, 2.2 Hz, H-6), 6.31 (1H, d, J = 2.2 Hz, H-8), 4.92 (1H, d, J = 11.8 Hz, H-2), 4.46 (1H, d, J = 11.8 Hz, H-3); 13C NMR (100 MHz, CD3OD): δC 193.1 (C-4), 165.4 (C-7), 163.6 (C-9), 145.6 (C-3′), 144.8 (C-4′), 128.7 (C-10), 128.6 (C-5), 119.5 (C-6′), 114.7 (C-2′), 114.5 (C-5′), 111.9 (C-10), 110.7 (C-6), 102.3 (C-8), 84.2 (C-2), 73.1 (C-3).26
(+)-Catechin (11) ‒ Colorless solid; C15H14O6; 1H NMR (400 MHz, CD3OD): δH 6.83 (1H, d, J = 1.9 Hz, H-2′), 6.75 (1H, d, J = 8.1 Hz, H-5′), 6.70 (1H, dd, J = 8.1, 1.9 Hz, H-6′), 5.93 (1H, d, J = 1.8, H-6), 5.86 (1H, d, J = 1.8 Hz, H-8), 4.56 (1H, d, J = 7.5 Hz, H-2), 3.97 (1H, dd, J = 7.5, 5.5 Hz, H-3), 2.83 (1H, dd, J = 16.1, 5.5 Hz, Ha-4), 2.50 (1H, dd, J = 16.1, 8.1 Hz, Hb-4); 13C NMR (100 MHz, CD3OD): δC 156.3 (C-7), 156.1 (C-5), 155.4 (C-9), 144.8 (C-4′), 144.8 (C-3′), 130.8 (C-1′), 118.7 (C-6′), 114.8 (C-5′), 114.8 (C-2′), 99.5 (C-10), 94.9 (C-6), 94.2 (C-8), 81.4 (C-2), 67.4 (C-3), 27.0 (C-4).27
Brazilin (12) ‒ Amorphous red powder; C16H14O5; 1H NMR (400 MHz, CD3OD): δH 7.17 (1H, d, J = 8.3 Hz, H-5), 6.69 (1H, s, H-2′), 6.58 (1H, s, H-5′), 6.45 (1H, dd, J = 8.3, 2.4 Hz H-6), 6.28 (1H, d, J = 2.4 Hz, H-8), 3.95 (1H, s, H-4), 3.91 (1H, d, J = 11.4 Hz, Ha-2), 3.67 (1H, d, J = 11.4 Hz, Hb-2), 3.00 (1H, d, J = 15.6 Hz, Ha-9), 2.75 (1H, d, J = 15.6 Hz, Hb-9); 13C NMR (100 MHz, CD3OD): δC 157.8 (C-7), 155.7 (C-8a), 145.6 (C-3′), 145.3 (C-4′), 137.4 (C-6′), 132.2 (C-5), 131.3 (C-1′), 115.5 (C-4a), 112.8 (C-2′), 112.4 (C-5′), 109.9 (C-6), 104.3 (C-8), 78.1 (C-3), 70.8 (C-2), 51.0 (C-4), 42.9 (C-9).28
3,4,5-Trimethoxyphenylβ-ᴅ-glucopyranoside (13) ‒ Amorphous white powder; C15H22O9; 1H NMR (400 MHz, DMSO-d6): δH 6.35 (2H, s, H-2, H-6), 4.74 (1H, d, J = 7.4 Hz, H-1′), 3.70 (6H, s, 3,5-OCH3), 3.55 (3H, s, 4-OCH3), 3.04‒3.41 (m, H-2′‒6′); 13C NMR (100 MHz, DMSO-d6): δC 154.4 (C-1), 153.5 (C-3, 5), 132.8 (C-4), 101.4 (C-1′), 94.7 (C-2, 6), 77.7 (C-5′), 77.2 (C-3′), 73.7 (C-2′), 70.5 (C-4′), 61.3 (C-6′), 60.5 (4-OCH3), 56.1 (3, 5-OCH3).29
1-(2-Methylbutyryl)phloroglucinol-glucopyranoside (14) ‒ Amorphous white powder; C17H24O9; 1H NMR (400 MHz, CD3OD): δH 6.16 (1H, d, J = 2.0 Hz, H-6), 5.94 (1H, d, J = 2.0 Hz, H-4), 5.02 (1H, d, J = 7.5 Hz, H-1′′), 3.88 (2H, m, H-2′, Ha-6′′), 3.70 (1H, dd, J = 12.1, 5.4 Hz, Hb-6′′), 3.35-3.50 (4H, m, H-2′′‒5′′), 1.79 (1H, dt, J = 14.2, 7.4 Hz, Hb-3′), 1.37 (1H, dt, J = 14.2, 7.4 Hz, Ha-3′), 1.11 (3H, d, J = 6.7 Hz, H-5′), 0.87 (3H, t, J = 7.4 Hz, H-4′); 13C NMR (100 MHz, CD3OD): δC 211.7 (C-1′), 167.4 (C-3), 165.8 (C-5), 161.7 (C-1), 106.7 (C-2), 101.6 (C-1′′), 98.3 (C-4), 95.4 (C-6), 78.7 (C-5′′), 78.4 (C-3′′), 74.8 (C-2′′), 71.2 (C-4′′), 62.5 (C-6′′), 46.9 (C-2′), 28.2 (C-3′), 16.8 (C-5′), 12.0 (C-4′).30
(+)-Epi-catechin (15) ‒ Colorless solid; C15H14O6; 1H NMR (400 MHz, CD3OD): δH 6.95 (1H, d, J = 1.9 Hz, H-2′), 6.75 (1H, dd, J = 8.1, 1.9 Hz, H-6′), 6.74 (1H, d, J = 8.1 Hz, H-5′), 5.92 (1H, d, J = 1.9, H-6), 5.90 (1H, d, J = 1.9 Hz, H-8), 4.79 (1H, s, H-2), 4.15 (1H, br s, H-3), 2.84 (1H, dd, J = 16.7, 4.5 Hz, Ha-4), 2.71 (1H, dd, J = 16.7, 2.8 Hz, Hb-4); 13C NMR (100 MHz, CD3OD): δC 156.5 (C-7), 156.2 (C-5), 155.9 (C-9), 144.5 (C-4′), 144.3 (C-3′), 130.8 (C-1′), 117.9 (C-6′), 114.5 (C-5′), 113.9 (C-2′), 99.6 (C-10), 94.9 (C-6), 94.5 (C-8), 78.4 (C-2), 66.0 (C-3), 27.8 (C-4).27
Astragalin (16) ‒Amorphous yellow powder; C21H20O11; 1H NMR (400 MHz, CD3OD): δH 8.07 (2H, d, J = 8.9 Hz, H-2′, 6′), 6.87 (2H, d, J = 8.9 Hz, H-3′, 5′), 6.39 (1H, d, J = 2.1 Hz, H-6), 6.19 (1H, d, J = 2.1 Hz, H-8), 5.13 (1H, d, J = 7.8 Hz, H-1′′), 3.42‒3.80 (6H, H-2′′‒6′′). 13C NMR (100 MHz, CD3OD): δC 179.7 (C-4), 166.1 (C-7), 163.1 (C-5), 161.6 (C-4′), 159.1 (C-2), 158.5 (C-9), 135.6 (C-3), 132.4 (C-2′, 6′), 122.7 (C-1′), 116.1 (C-3′, 5′), 105.7 (C-10), 104.9 (C-1′′), 99.9 (C-6), 94.8 (C-8), 77.1 (C-3′′), 75.0 (C-5′′), 73.0 (C-2′′), 70.0 (C-4′′), 61.9 (C-6′′).31
Mixture of two conformers of protosappanin B (17/18) ‒ Colorless solid; C16H16O6.
Major form of protosappanin B (17): 1H NMR (400 MHz, CD3OD): δH 6.96 (d, J = 8.0 Hz, H-1), 6.72 (ovl, H-9, 12), 6.53 (ovl, H-2), 6.44 (s, H-4), 4.14 (d, J = 12.4 Hz, Ha-6), 3.85 (d, J = 12.4 Hz, Hb-6), 3.53 (ovl, Ha-7a), 3.45 (d, J = 11.2 Hz, Hb-7a), 2.54 (d, J = 13.6 Hz, Ha-8), 2.48 (d, J = 13.6 Hz, Hb-8). 13C NMR (100 MHz, CD3OD): δC 159.3 (C-4a), 158.9 (C-3), 144.9 (C-10, 11), 133.2 (C-1), 132.5 (C-12a), 127.5 (C-8a), 124.1 (C-12b), 119.9 (C-9), 117.5 (C-12), 111.5 (C-2), 108.2 (C-4), 76.7 (C-6), 73.0 (C-7), 68.4 (C-7a), 39.9 (C-8).32
Minor form of protosappanin B (18): 1H NMR (400 MHz, CD3OD): δH 6.96 (d, J = 8.0 Hz, H-1), 6.72 (ovl, H-9), 6.66 (s, H-12), 6.53 (ovl, H-2), 6.50 (s, H-4), 4.37 (d, J = 12.0 Hz, Ha-6), 3.53 (ovl, Hb-6, Ha-7a), 3.39 (d, J = 11.2 Hz, Hb-7a), 2.67 (s, H-8). 13C NMR (100 MHz, CD3OD): δC 160.4 (C-4a), 159.0 (C-3), 144.8 (C-10), 144.8 (C-11), 132.7 (C-1), 132.0 (C-12a), 128.1 (C-8a), 125.2 (C-12b), 119.0 (C-9), 117.9 (C-12), 112.1 (C-2), 108.8 (C-4), 76.7 (C-6), 73.3 (C-7), 65.7 (C-7a), 42.7 (C-8).32
α-Glucosidase inhibition assay - α-Glucosidase inhibitory assay was conducted spectrophotometrically using p-nitrophenyl α-ᴅ-glucopyranoside (p-NPG) as the substrate, according to the previously published procedure with a slight modification.33 In brief, 20 μL enzyme α-glucosidase (0.1 unit) was added to 96 well plates, which contained 20 μL samples and 20 μL phosphate buffer (100 mM, pH = 6.8). Subsequently, the enzyme reaction was started by the addition of 20 μL p-NPG (2.5 mM) and then incubated for 10 min at 37oC. After 10 min, 80 μL Na2CO3 0.2 M was used for each well to finalize the enzyme reaction. The absorbance was measured at 405 nm using a microplate spectrophotometer. Acarbose was used as a positive control. The percentage inhibition (%) is calculated using the following equation:
In which Ac and As are the average value of control and sample, respectively.
Enzyme kinetics of α-glucosidase inhibition - To determine the mode of α-glucosidase inhibition of the most active compounds (1, 6, 7, and 9), two kinetic methods including Lineweaver-Burk plots and Dixon plots were used.33 The α-glucosidase inhibition was monitored at various concentrations of substrate p-NPG (0.625, 1.25, and 2.5 mM) and different concentrations of tested compounds (1.71, 4.44, and 8.41 μM for 1; 3.0, 7.0, and 25.0 μM for 6; 8.0, 16.0, and 26.0 μM for 7; 5.0, 10.0, and 25.0 μM for 9). The enzyme reaction was carried out as described above. The obtained results were analyzed by using SigmaPlot 12.0 software (SPCC Inc, Chicago IL, U.S.A). Kinetic constant values (Ki) were calculated due to the Dixon plots.
Statistical analysis ‒ All experimental bioassay data were performed as the mean ± standard error (SE) of the mean from at least three independent experiments. One-way analysis of variance (ANOVA) and Dunnett’s test were applied to evaluate statistical significance. P values of less than 0.05 were considered to be statistically significant.
Result and Discussion
Phytochemical investigation of C. decapetala var. japonica stems resulted in the isolation of 16 known compounds (1‒16) (Fig. 1), which were identified as quercetin (1),17 methyl gallate (2),18 caesalpiniaphenol C (3),19 8S,8′S,7′R-(‒)-lyoniresinol (4),20 7,3′,5′-trihydroxyflavanone (5),21 sappanchalcone (6),22 sappanone A (7),34 taxifolin (8),24 fisetin (9),25 fustin (10),26 (+)-catechin (11),27 brazilin (12),28 3,4,5-trimethoxyphenyl β-ᴅ-glucopyranoside (13),29 1-(2-methylbutyryl)phloroglucinol-glucopyranoside (14),30 (+)-epi-catechin (15),27 and astragalin (16)31, and a mixture of two conformers of protosappanin B (17/18),32 by analyzing and comparing their spectroscopic data with those reported in the literature. Among them, caesalpiniaphenol C (3), 8S,8′S,7′R- (‒)-lyoniresinol (4), taxifolin (8), fisetin (9), and fustin (10) were isolated from C. decapetala for the first time. In addition, this represents the first isolation of 3,4,5-trimethoxyphenyl β-ᴅ-glucopyranoside (13) and 1-(2-methylbutyryl) phloroglucinol-glucopyranoside (14) from the Caesalpinia genus.
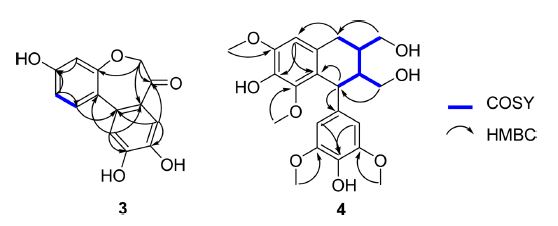
Compound 3 was obtained as a yellow needle. The 1H and 13C NMR spectra of 3 showed one aromatic ABX system at [δH/δC 7.06 (1H, d, J = 8.4 Hz, H-1)/130.9 (C-1), 6.66 (1H, dd, J = 8.4, 1.8 Hz, H-2)/113.3 (C-2), and 6.62 (1H, d, J = 1.8 Hz, H-4)/108.8 (C-4)], one 1,2,4,5-tetrasubstituted benzene ring assigned at δH/δC 6.68 (2H, s, H-8, 11)/117.4 (C-8, 11). It also displayed one methylene group at δH/δC 4.42 (2H, s, H2-6)/78.8 (C-6). Besides, the 13C NMR spectrum of 3 exhibited one carbonyl carbon signal at δC 207.7 (C-7). The NMR data of 3 closely resembled those of caesalpiniaphenol C reported in the literature.19 The complete structure of 3 was further determined by the key HMBC correlations of H-6/C-4a, H-6/C-7, H-6/C-7a, H-8/C-7, H-1/C-11a, H-11/C-11a, H-2/C-11b, and H-11/C-11b (Fig. 2). Therefore, 3 was determined to be caesalpiniaphenol C.19
Compound 4 was obtained as an amorphous, white powder with a negative optical rotation [ ‒40 (c = 0.025, MeOH)]. The 1H and 13C NMR spectra of 4 displayed typical signals for a 4-aryltetralin type lignan, which revealed one 1,2,3,4,5-pentasubstituted benzene ring at [δH/δC 6.56 (1H, s, H-2)/106.3 (C-2)], one 1,3,4,5-tetrasubstituted ring at [δH/δC 6.36 (2H, s, H-2′, 6′)/105.4 (C-2′, 6′)], four aromatic methoxy groups at [δH/δC 3.83 (3H, s, 3-OCH3)/55.3 (3-OCH3), 3.35 (3H, s, 5-OCH3)/58.7 (5-OCH3), 3.71 (6H, s, 3′, 5′-OCH3)/55.2 (3′, 5′-OCH3)], two oxygenated methylene groups at [δH/δC 3.48 (2H, m, Ha-9, Ha-9′), 3.57 (1H, dd, J = 10.8, 5.0 Hz, Hb-9), 3.64 (1H, d, J = 5.5 Hz, Hb-9′)/65.3 (C-9), 62.7 (C-9′)], one methylene signal at [δH/δC 2.55 (1H, dd, J = 15.0, 11.8 Hz, Ha-7), 2.68 (1H, dd, J = 15.0, 4.8 Hz, Hb-7)/32.1 (C-7)], and three methine groups at [δH/δC 4.29 (1H, d, J = 5.7 Hz, H-7′)/40.9 (C-7′), 1.95 (1H, dd, J = 8.1, 5.5 Hz, H-8′)/47.6 (C-8′), 1.61 (1H, m, H-8)/39.4 (C-8)]. Based on the above 1D NMR data together with the analysis of the HMQC and HMBC data (Fig. 2), 4 was determined to be lyoniresinol.20 Consequently, the absolute configuration of 4 was identified as 8S,8′S,7′R-(‒)-lyoniresinol based on these observations and comparison with its reported literature.20
Compound 8 was obtained as an amorphous colorless powder. The 1H and 13C NMR spectra of 8 showed resonances characteristic of a flavanonol with signals of one ABX coupling system at [δH 6.95 (1H, d, J = 1.5 Hz, H-2′), 6.78 (1H, d, J = 8.1 Hz, H-5′), 6.83 (1H, dd, J = 8.1, 1.5 Hz, H-6′)], one 1,2,3,5-tetrasubstituted ring, which was supported by two meta coupling protons at [δH 5.86 (1H, d, J = 1.5 Hz, H-6), 5.90 (1H, d, J = 1.5 Hz, H-8)], two oxygenated methine protons at δH 4.87 (1H, d, J = 11.5 Hz, H-2), 4.48 (1H, d, J = 11.5 Hz, H-3), and one carbonyl carbon signal at δC 196.9 (C-4). The 13C NMR spectrum of 8 revealed 15 carbon signals corresponding to the predicted structure. Thus, the chemical structure of 8 was established to be taxifolin.24
Compound 9 was obtained as an amorphous white powder. The 1H NMR spectrum of 9 displayed signals of two ABX coupling systems at δH 7.96 (1H, dd, J = 9.4, 0.5 Hz, H-5), 6.88 (3H, m, H-6, H-8, H-5′), 7.75 (1H, d, J = 2.1 Hz, H-2′), 7.65 (1H, ddd, J = 8.5, 2.1, 0.5 Hz, H-6′). Its 13C NMR spectrum exhibited 15 carbon signals, including 12 carbon signals for two substituted benzene rings at δC 126.0 (C-5), 114.5 (C-6), 162.9 (C-7), 101.5 (C-8), 157.1 (C-9), 113.9 (C-10), 122.9 (C-1′), 114.5 (C-2′), 146.0 (C-3′), 147.2 (C-4′), 114.8 (C-5′), 120.2 (C-6′), two oxygenated olefinic carbon signals at δC 144.8 (C-2), and 137.1 (C-3), and one carbonyl signal at δC 172.9 (C-4). Based on a comparison of the above 1D NMR data with those in the literature,25 9 was determined as a flavonol (fisetin).
Compound 10 was obtained as a white needle. The proton and carbon spectra of 10 were similar to those of 8 except for the signals belonging to ring A with one ABX coupling system at [δH/δC 7.69 (1H, d, J = 8.7 Hz, H-5)/128.6 (C-5), 6.50 (1H, dd, J = 8.7, 2.2 Hz, H-6)/110.7 (C-6), 6.31 (1H, d, J = 2.2 Hz, H-8)/102.3 (C-8)], in 10. In the comparison with the previous report, 10 was identified to be fustin.26
Compound 13 was obtained as a white powder. The proton spectrum of 13 showed two singlet proton signals for two symmetric protons belonging to one 1,3,4,5-tetrasubstituted benzene ring at δH 6.35 (2H, s, H-2, H-6), and three aromatic methoxy groups at δH 3.70 (6H, s, 3, 5-OCH3), and 3.55 (3H, s, 4-OCH3). In addition, an anomeric proton signal with a large J coupling constant was observed at δH 4.74 (1H, d, J = 7.4 Hz, H-1′). The β-glucose moiety was further confirmed by the 13C NMR spectrum with 6 typical carbon signals at δC 101.4 (C-1′), 73.7 (C-2′), 77.2 (C-3′), 70.5 (C-4′), 77.7 (C-5′), and 61.3 (C-6′). The position of glucose moiety was located at C-1 (δC 154.4) due to the HMBC correlation between anomeric proton H-1′ and C-1. Therefore, 13 was established as 3,4,5-trimethoxyphenyl β-ᴅ-glucopyranoside.29
Compound 14 was obtained as an amorphous white powder. The 1H NMR spectrum of 14 showed the aliphatic signals for a 2-methylbutyryl moiety at [δH 1.37 (1H, dt, J = 14.2, 7.4 Hz, Ha-3′), 1.79 (1H, dt, J = 14.2, 7.4 Hz, Hb-3′), 0.87 (3H, t, J = 7.4 Hz, H-4′), 1.11 (3H, d, J = 6.7 Hz, H-5′)],30 and an anomeric proton at δH 5.02 (1H, d, J = 7.5 Hz, H-1′′). Besides, two meta-coupled doublets appeared at δH 5.94 (1H, d, J = 2.0 Hz, H-4), 6.16 (1H, d, J = 2.0 Hz, H-6), indicating an asymmetrically substituted ring. The glucose moiety was attached to δC 161.7 (C-1), which was confirmed by the HMBC cross-peaks between H-1′′ and C-1. Therefore, 14 was determined to be 1-(2-methylbutyryl)phloroglucinol-glucopyranoside.30
The isolated compounds were evaluated for their inhibitory effect against α-glucosidase enzyme with acarbose was used as a positive control (IC50 = 152.22 ± 1.86 μM). Our results showed that the isolated flavonols, quercetin (1), and fisetin (9) exhibited a potent inhibitory effect, presenting IC50 values of 5.80 ± 0.25 μM and 10.82 ± 0.19 μM, respectively, which were much lower than acarbose (Fig. 3). While astragalin (16), a flavonol glycoside suppressed slightly α-glucosidase enzyme action (IC50 > 50 μM) (Table 1). The chalcone 6 (sappanchalcone) and homoisoflavanone 7 (sappanone A) showed a significant α-glucosidase inhibitory activity with IC50 values of 15.01 ± 0.34 μM and 14.13 ± 0.27 μM, respectively. The isolated flavanone 5 (7,3′,5′-trihydroxyflavanone) and flavan-3-ol 15 (epi-catechin) exhibited an inhibitory effect of α-glucosidase with IC50 values of 22.56 ± 0.54 μM and 30.85 ± 0.40 μM, respectively. Besides, caesalpiniaphenol C (3), and the mixture (17/18) also suppressed α-glucosidase action with IC50 values of 34.82 ± 0.25 μM and 49.51 ± 0.06 μM, respectively. The remaining compounds (2‒4, 8, 10‒14) exhibited a weak effect on the inhibition of α-glucosidase (IC50 > 50 μM) (Table 1).
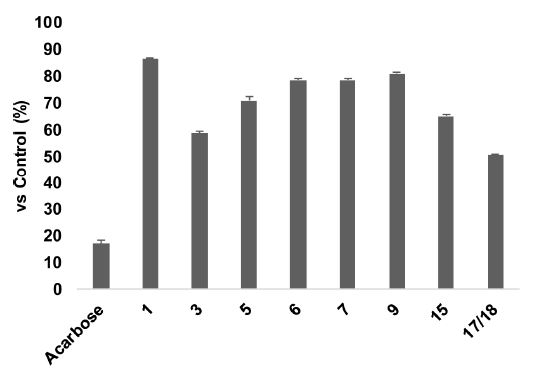
Inhibitory effects (%) of the active compounds (1, 3, 5, 6, 7, 9, 15, and 17/18) and acarbose on the enzymatic activities of α-glucosidase at a concentration of 50 μM. The data are expressed as mean ± SE. The experiments were done in triplicates.
For the potent inhibitors (1, 6, 7, and 9), we continuously examined their inhibition kinetics in vitro. α-Glucosidase was treated individually with four concentrations of tested compounds, using p-NPG as a substrate (0.625 ‒ 2.5 mM). The type of inhibition and inhibition constants (Ki values) of the above compounds was deduced from two kinetic methods: Lineweaver-Buck plots and Dixon plots (Fig. 4). After analysis of these two types of graphs and based on the best fit results and goodness to fit analysis, we concluded that two flavonols (1 and 9) (Fig. 4A and 4D) showed a non-competitive inhibition type with Ki values of 5.8 and 10.8 μM, respectively, while chalcone (6) and homoisoflavanone (7) (Fig. 4B and 4C) had shown a mixed inhibition type with Ki values of 11.8 and 8.0 μM, respectively. The Ki values were calculated using Dixon plot by plotting the reciprocal of maximum velocity (1/Vmax) (y-axis) against the concentrations (x-axis) (Fig. 4E‒H).
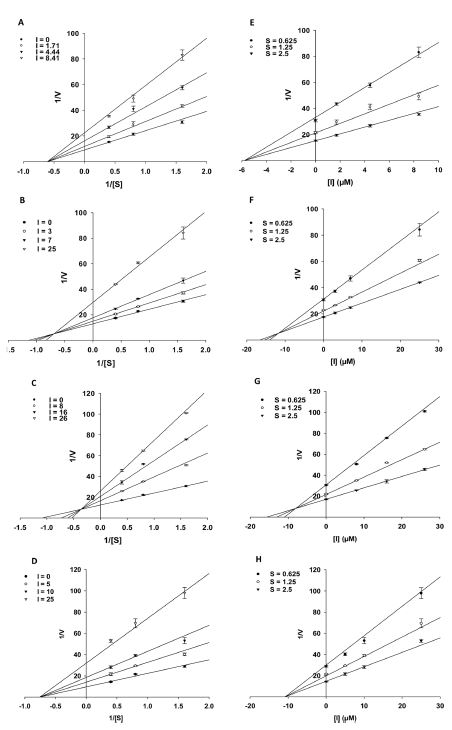
The inhibition kinetics of compounds 1, 6, 7, and 9 towards α-glucosidase. (A‒D) Lineweaver-Burk plots for α-glucosidase inhibition of compounds 1 (A), 6 (B), 7 (C), and 9 (D). (E‒H) Dixon plots for α-glucosidase inhibition of compounds 1 (E), 6 (F), 7 (G), and 9 (H).
Interestingly, two flavonols (1 and 9), which contained one carbonyl carbon and one double bond at the C-ring inhibited strongly α-glucosidase enzyme action. The absence of both these characteristics at the C-ring in the case of flavan-3-ols (11 and 15), or the lack of a double bond at C-2 in the C ring in the case of flavanone (5) and flavanonols (8 and 10) induced a significant reduction in the level of inhibition. Meanwhile, the addition of one glucose moiety at C-3 in the structure of 16 also decreased dramatically its inhibitory activity. This evidence supported the key role of flavones in anti-diabetic drug research. Our findings for α-glucosidase inhibitory activity of compounds 1, 5, 6, 9, 15‒18 were in accordance with those previously reported.35,36 On the other hand, this is the first report on inhibiting α-glucosidase action for caesalpiniaphenol C (3) and sappanone A (7). Furthermore, the α-glucosidase enzyme kinetic inhibition types of sappanchalcone (6) and sappanone A (7) were also investigated for the first time.
In summary, sixteen known phenolic compounds and one known mixture were isolated from the C. decapetala var. japonica stems. Five compounds (3, 4, 8, 9, and 10) were reported from this plant for the first time, and two compounds (13 and 14) were found in this genus for the first time. Our research concluded that C. decapetala var. japonica and its flavonoids, especially flavonols, homoisoflavanone, and chalcones were considered to be alternative inhibitors for the regulation of hyperglycemia.
Acknowledgments
This study was supported by the National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT (2021R1A2C2011940), Korea. We are thankful to the Korea Basic Science Institute (KBSI) for mass spectrometric measurements.
References
-
Lago, R. M.; Singh, P. P.; Nesto, R. W. Nat. Clin. Pract. Endocrinol. Metab. 2007, 3, 667.
[https://doi.org/10.1038/ncpendmet0638]

- Van de Laar, F. A.; Lucassen, P. L. B. J.; Akkermans, R. P.; Van de Lisdonk, E. H.; Rutten, G. E. H. M.; Van Weel, C. Cochrane Database Syst. Rev. 2005. CD003639.
-
Poovitha, S.; Parani, M. BMC Complement Altern. Med. 2016, 16, 185.
[https://doi.org/10.1186/s12906-016-1085-1]

-
Tu, W. C.; Ding, L. F.; Peng, L. Y.; Song, L. D.; Wu, X. D.; Zhao, Q. S. Phytochemistry 2022, 193, 112973.
[https://doi.org/10.1016/j.phytochem.2021.112973]

- Shah, G. M.; Jamal, Z.; Hussain, M. Pak. J. Bot. 2013, 45, 253-261.
-
Chaudhary, C. M.; Jan, H. A.; Kunwar, R. M.; Bussmann, R. W.; Paniagua-Zambrana, N. Y. In Ethnobotany of the Himalayas: Caesalpinia decapetala (Roith) Alston Fabaceae; Bussmann, R. W.; Paniagua-Zambrana, N. Y. Ed; Springer International Publishing; 2020, pp 1-8.
[https://doi.org/10.1007/978-3-030-45597-2_43-1]

-
Miyazawa, M.; Nagata, T.; Nakahashi, H.; Takahashi, T. J. Essent. Oil Res. 2012, 24, 441-446.
[https://doi.org/10.1080/10412905.2012.703475]

- Parveen, A.; Akash, M. S. H.; Rehman, K.; Mahmood, Q.; Qadir, M. I. BioImpacts. BI 2014, 4, 43-48.
-
Wei, X. H.; Yang, S. J.; Liang, N.; Hu, D. Y.; Jin, L. H.; Xue, W.; Yang, S. Molecules 2013, 18, 1325-1336.
[https://doi.org/10.3390/molecules18011325]

-
Kamikawa, S.; Oshimo, S.; Ohta, E.; Nehira, T.; Ômura, H.; Ohta, S. Phytochemistry 2016, 121, 50-57.
[https://doi.org/10.1016/j.phytochem.2015.10.001]

-
Zhang, L.; Chen, J.; Ke, C.; Zhang, H.; Zhang, S.; Tang, W.; Liu, C.; Liu, G.; Chen, S.; Hu, A.; Sun, W.; Xiao, Y.; Liu, M.; Chen, X. Viruses 2020, 12, 557.
[https://doi.org/10.3390/v12050557]

-
Ogawa, K.; Aoki, I.; Sashida, Y. Phytochemistry 1992, 31, 2897-2898.
[https://doi.org/10.1016/0031-9422(92)83658-L]

-
Van Kiem, P.; Van Minh, C.; Huong, H. T.; Lee, J. J.; Kim, Y. H. Chem. Pharm. Bull(Tokyo). 2005, 53, 428-430.
[https://doi.org/10.1248/cpb.53.428]

-
Wei, X. H.; Yang, S. J.; Liang, N.; Hu, D. Y.; Jin, L. H.; Xue, W.; Yang, S. Molecules 2013, 18, 1325-1336.
[https://doi.org/10.3390/molecules18011325]

-
Hussain, L.; Qadir, M. I.; Rehman, S. Bangladesh J. Pharmacol. 2014, 9, 529-532.
[https://doi.org/10.3329/bjp.v9i4.19961]

- Shelar, U. B. Phytochemical And Pharmacological Study of Caesalpinia Decapetala Leaves; M. K. Infoedutech Private Limited: India, 2022. pp 1-171.
-
Charisiadis, P.; Kontogianni, V. G.; Tsiafoulis, C. G.; Tzakos, A. G.; Siskos, M.; Gerothanassis, I. P. Molecules 2014, 19, 13643-13682.
[https://doi.org/10.3390/molecules190913643]

-
Hernández-García, E.; García, A.; Avalos-Alanís, F. G.; Rivas-Galindo, V. M.; Delgadillo-Puga, C.; Rayo Camacho-Corona, M. D. Data Brief 2018, 22, 255-268.
[https://doi.org/10.1016/j.dib.2018.12.008]

-
Cuong, T. D.; Hung, T. M.; Kim, J. C.; Kim, E. H.; Woo, M. H.; Choi, J. S.; Lee, J. H.; Min, B. S. J. Nat. Prod. 2012, 75, 2069-2075.
[https://doi.org/10.1021/np3003673]

-
Cretin, B. N.; Sallembien, Q.; Sindt, L.; Daugey, N.; Buffeteau, T.; Waffo-Teguo, P.; Dubourdieu, D.; Marchal, A. Anal. Chim. Acta 2015, 888, 191-198.
[https://doi.org/10.1016/j.aca.2015.06.061]

- Fu, J.; Liang, G. Y.; Zhang, J. X.; Zhang, Y. P.; Liu, L.; Pan, W. D.; Cao, P. X. Xiandai Yaowu Yu Linchuang 2013, 28, 265-268.
-
Jung, E. G.; Han, K. I.; Kwon, H. J.; Patnaik, B. B.; Kim, W. J.; Hur, G. M.; Nam, K. W.; Han, M. D. Arch. Pharm. Res. 2015, 38, 973-983.
[https://doi.org/10.1007/s12272-015-0557-z]

-
Chang, T. S.; Chao, S. Y.; Ding, H. Y. Int. J. Mol. Sci. 2012, 13, 10359-10367.
[https://doi.org/10.3390/ijms130810359]

-
Khlupova, M.; Vasil'eva, I.; Shumakovich, G.; Morozova, O.; Chertkov, V.; Shestakova, A.; Kisin, A.; Yaropolov, A. J. Mol. Catal. B Enzym. 2016, 123, 62-66.
[https://doi.org/10.1016/j.molcatb.2015.11.010]

-
Kim, J. S.; Kwon, Y. S.; Chun, W. J.; Kim, T. Y.; Sun, J.; Yu, C. Y.; Kim, M. J., Food Chem. 2010, 120, 539-543.
[https://doi.org/10.1016/j.foodchem.2009.10.051]

- Kim, M. Y.; Chung, I. M.; Choi, D. C.; Park, H. J., Nat. Prod. Sci. 2009, 15, 208-212.
- Abd El-Razek, M. H. Asian J. Chem. 2007, 19, 4867.
-
Namikoshi, M.; Nakata, H.; Yamada, H.; Nagai, M.; Saitoh, T. Chem. Pharm. Bull. 1987, 35, 2761-2773.
[https://doi.org/10.1248/cpb.35.2761]

-
Verotta, L.; Dell'Agli, M.; Giolito, A.; Guerrini, M.; Cabalion, P.; Bosisio, E. J. Nat. Prod. 2001, 64, 603-607.
[https://doi.org/10.1021/np000306j]

-
Bohr, G.; Gerhäuser, C.; Knauft, J.; Zapp, J.; Becker, H. J. Nat. Prod. 2005, 68, 1545-1548.
[https://doi.org/10.1021/np050164z]

- Fawzi, F.; Mahdi, M. F.; Abaas, I. S. J. Pharm. Sci. 2019, 11, 185-190.
-
Zhao, M. B.; Cai, C. Q.; Tu, P. F.; Tang, L. Magn. Reson. Chem. 2016, 54, 601-605.
[https://doi.org/10.1002/mrc.4408]

-
Ha, M. T.; Lee, T. H.; Kim, C. S.; Prajapati, R.; Kim, J. A.; Choi, J. S.; Min, B. S. Phytochemistry 2022, 197, 113100.
[https://doi.org/10.1016/j.phytochem.2022.113100]

-
Lee, S.; Choi, S. Y.; Choo, Y. Y.; Kim, O.; Tran, P. T.; Dao, C. T.; Min, B. S.; Lee, J. H. Int. Immunopharmacol. 2015, 28, 328-336.
[https://doi.org/10.1016/j.intimp.2015.06.015]

-
Proença, C.; Freitas, M.; Ribeiro, D.; Oliveira, E. F. T.; Sousa, J. L. C.; Tomé, S. M.; Ramos, M. J.; Silva, A. M. S.; Fernandes, P. A.; Fernandes, E. J. Enzyme Inhib. Med. Chem. 2017, 32, 1216-1228.
[https://doi.org/10.1080/14756366.2017.1368503]

-
Arsiningtyas, I.; Kato, E.; Kawabata, J. Planta Med. 2015, 81, 42.
[https://doi.org/10.1055/s-0035-1565419]