Enzyme Kinetics and Molecular Docking Investigation of Acetylcholinesterase and Butyrylcholinesterase Inhibitors from the Marine Alga Ecklonia cava
Abstract
Ecklonia cava Kjellman (Laminareaceae) grows along the coast of Jeju Island, Korea, and is well-known in Korea for its use as a food ingredient, animal feed, and medicine. This seaweed contains phlorotannins, polymerized units of phloroglucinol, a term derived from phloroglucinol, the building block of these complex molecules, which is also the common name for these compounds. Phlorotannins are secondary metabolites that hold significance for human health due to their various beneficial properties, including antioxidant, anti-cancer, anti-allergy, and anti-HIV activities. In this study, 10 phlorotannins (1–10) were isolated from an 80% EtOH extract of E. cava. The structures of these compounds were determined through spectroscopic analysis and comparison with the literature. The inhibitory effects of compounds 1–10 on acetylcholinesterase (AChE) and butyrylcholinesterase (BuChE) were investigated. In an AChE inhibition assay, compounds 1, 2, 4, and 6–10 had IC50 values ranging from 0.9 ± 0.8 to 66.5 ± 0.4 µM; compounds 4, 6, and 9 had potent BuChE inhibitory effects, with IC50 values ranging from 1.4 ± 3.8 to 25.2 ± 0.1 µM. Furthermore, enzyme kinetics and molecular docking simulations were conducted to gain insights into the inhibition mode, binding mechanism, and crucial interactions between these active compounds and the target enzyme. This indicates that E. cava is a potentially valuable natural source of AChE and BuChE inhibitors.
Keywords:
Ecklonia cava, Laminareaceae, phlorotannin, AChE, BuChEIntroduction
Alzheimer’s disease (AD) is a chronic progressive neurodegenerative disorder characterized by progressive cognitive decline, behavioral disturbances, and challenges in daily living activities.1 The classical neurodegenerative pathological changes in AD in the brain include the formation of β-amyloid plaques, neurofibrillary tangles, neuronal cell death, and significant synaptic loss.2,3 However, the precise cause of AD is not known, although it is linked to substantial early reductions in presynaptic markers of the cholinergic system.4 The synthesis of cholinergic acetyltransferase and acetylcholine (ACh) decreases as cholinergic neurons are lost, leading to a gradual decline in cholinergic neurotransmission with disease progression.4 Cholinesterase (ChE) inhibitors disrupt the breakdown of ACh following synaptic release and constitute a primary treatment for AD.5,6 Recently, the glutamatergic antagonist memantine was introduced as a treatment option.7 While ChE inhibitors consistently offer cognitive improvements in mild to moderate AD, their effects are generally modest.8,9 The clinical relevance of these drugs is under ongoing discussion. Until recently, the role of butyrylcholinesterase (BuChE) in regulating ACh levels had been overlooked.10,11 However, a growing body of evidence suggests that both acetylcholinesterase (AChE) and BuChE play significant roles in regulating ACh levels and may contribute to the development and progression of AD.10–12 Selective BuChE inhibition might confer cognitive benefits similar to classical AChE inhibition without the characteristic adverse effects that often come with higher doses.10,13
Marine algae have a rich history of traditional use as food and folk medicine in Asia.14 These algae are abundant sources of bioactive compounds that vary in structure, holding significant potential for pharmaceutical and biomedical applications. Ecklonia cava Kjellman, a member of the Laminariaceae family, thrives off the coast of Jeju Island, Korea.15 This species is widely embraced in Korea for its utility as a culinary ingredient, animal feed, and traditional remedy.16 In 2008, a commercial extract derived from E. cava was approved as a new dietary ingredient by the U.S. Food and Drug Administration (FDA) (FDA-1995-S-0039-0176).17,18
Phlorotannins, distinct polyphenolic compounds found in E. cava and other brown algae, have garnered increasing attention due to their remarkable biological properties and potential therapeutic applications.19,20 These compounds have complex chemical structures characterized by the polymerization of phloroglucinol units, resulting in diverse forms and sizes.21 This structural complexity contributes to their variable and potent bioactivities. One of the most prominent characteristics of phlorotannins is their potent antioxidant activity; they can neutralize harmful free radicals, which are implicated in oxidative stress and various chronic diseases.19,22 By safeguarding cells and tissues from oxidative damage, phlorotannins contribute to the overall maintenance of health and may play a role in reducing age-related conditions.21,23,24 They also have various pharmacological effects, including anti-cancer, anti-allergy, and anti-HIV activities.22,25–29
As part of our ongoing research into the phytochemical components of Korean medicinal plants with potential applications in AD treatment, in this study, we successfully isolated 10 phlorotannins (1–10) from an extract of E. cava. The structures of these compounds were elucidated through a combination of spectroscopic analysis and literature comparisons. Their inhibitory effects on AChE and BuChE were evaluated. Furthermore, insights into the inhibition mode, binding mechanism, and key interactions of active compounds with the target enzyme were gained by performing in vitro enzyme kinetic analysis and in silico docking simulations.
Experimental
General experimental procedures – The solvents used for material separation, including MeOH, n-hexane, CHCl3, CH2Cl2, EtOAc, n-BuOH, were purchased from SK Chemical (Suwon, Korea). Chromatographic seperations were carried out by thin layer chromatography (TLC). The TLC was carried out glass plates pre-coated with silica gel 60 F254 and RP-18 F254 (20 × 20 cm, Merck, Darmstadt, Germany) and visualized under UV at 254 nm and 365 nm. TLC color reagent was used 10% ethanol-sulfuric acid. Column chromatography (CC) was conducted using 230-400 mesh silica gel (Kieselgel 60, Merck) and RP-18 (ODS-A, 12 nm, S-150 mM, YMC, Tokyo, Japan). Nuclear magnetic resonance (NMR) experiments were conducted on Bruker Fourier 300 (1H-300 MHz, 13C-75 MHz) and Jeol JNM-AL 400 (1H-400 MHz, 13C-100 MHz) spectrometers. In the inhibition assay, K2HPO4 and KH2PO4 were purchased from Daejung Chemical. AChE (derived from Electrophorus electricus, C3389), BuChE (derived from Equine serum, C7512), 9-amino-1,2,3,4-tetrahydroacridine hydrochloride hydrate (tacrine, A79922-5G), acetylthiocholine iodide (A5751-5G), butyrylthiocholine iodide (B3253-5G), and 5,5′-dithiobis (2-nitrobenzoic acid) (DTNB, D8130-5G) were purchased from Sigma Aldrich (MO, USA). Absorbance intensity measurement was conducted using Tecan Infinite M200 microplate reader (Zürich, Switzerland).
Plant material – Dried E. cava was provided by Uniwell (1251, Garosu-ro, Heungdeok-gu, Cheongju-si, Chungcheongbuk-do, Korea, 2015). The plant authentication was verified by one of the authors (Prof. Young Ho Kim). A voucher specimen (CNU-15005) has been deposited at the Laboratory of Pharmacognosy, College of Pharmacy, Chungnam National University, Daejeon, Korea.
Extraction and isolation – The dried powder (1.0 kg) of E. cava was refluxed with 80% EtOH 16 L for 72 h, and the ethanol extract was concentrated in vacuo to obtain with 80% EtOH extract (290.0 g). The extract was suspended in water (2.0 L), and the aqueous layer was partitioned with n-hexane, ethyl acetate (EA), and n-BuOH. The EA fraction (54.9 g) was subjected to silica gel CC eluted with CH2Cl2:MeOH (gradient 8:1 → 2:1, v/v) to obtain four fractions (1A – 1D). Fraction 1B was chromatographed on silica gel CC eluting with CHCl3: MeOH:H2O (7:1:0.1 → 1:1:0.1, v/v/v) to afford six fractions (2A – 2F). Fraction 2C was chromatographed on RP-18 CC eluting with acetone:H2O (1:3, v/v) to isolate compounds 1 (234.2 mg) and 3 (25.1 mg). Fraction 2D was subjected to RP-18 CC eluted with acetone:H2O (1:3, v/v) to obtain compound 2 (128.5 mg). The fraction 2E was subjected to RP-18 column using MeOH:H2O (1:3, v/v) to isolate compound 4 (230.2 mg) and five subfractions (3A – 3E). Subfraction 3A was chromatographed on RP-18 CC, using MeOH:H2O (1:3, v/v) as eluent, to isolate compound 5 (304.4 mg). Subfraction 3E was chromatographed on RP-18 column eluting with acetone: MeOH:H2O (1:1:3, v/v/v) to obtain compounds 6 (85.5 mg) and 7 (69.0 mg). Fraction 2F was subjected to RP-18 CC, using MeOH:H2O (1:4, v/v) as mobile phase, to obtain compound 8 (52.7 mg). Fraction 1C was subjected to RP-18 CC eluting with acetone:H2O (1:5, v/v) to obtain compound 9 (110.5 mg). Fraction 1D was chromatographed on RP-18 CC eluting with acetone:H2O (1:4, v/v) to isolate compound 10 (67.5 mg).
Phloroglucinol (1) – White powder; ESI-MS: m/z 125 [M−H]−; 1H-NMR (CD3OD, 300 MHz): δH 5.71 (3H, s, H-2, H-4, H-6); 13C-NMR (CD3OD, 75 MHz): δC 160.0 (C-1, C-3, C-5), 95.5 (C-2, C-4, C-6).30
Eckol (2) – Light brown powder; ESI-MS: m/z 371 [M−H]−; 1H and 13C-NMR data, see reference.31,32
Dioxinodehydroeckol (3) – White powder; ESI-MS: m/z 369 [M−H]−; 1H-NMR (DMSO-d6, 300 MHz): δH 9.80 (1H, s, OH-1), 9.68 (1H, s, OH-9), 9.65 (1H, s, OH-6), 9.32 (1H, s, OH-3), 9.31 (1H, s, OH-11), 6.14 (1H, s, H-7), 6.06 (1H, d, J = 2.7 Hz, H-2), 6.02 (1H, d, J = 2.7 Hz, H-10), 5.82 (1H, d, J = 2.7 Hz, H-4), 5.80 (1H, d, J = 2.7 Hz, H-12); 13C-NMR (DMSO-d6, 75 MHz): δC 153.4 (C-3), 153.1 (C-11), 146.2 (C-1), 146.0 (C-9), 142.0 (C-4a), 141.7 (C-12a), 140.2 (C-6), 137.2 (C-7a), 131.6 (C-13b), 125.9 (C-5a), 122.6 (C-8a), 122.4 (C-13a), 122.2 (C-14a), 98.9 (C-2, C-10), 97.6 (C-7), 93.9 (C-4, C-12).31
Phlorofucofuroeckol A (4) – Amorphous powder; ESI-MS: m/z 601 [M−H]−; 1H and 13C-NMR data, see reference.31
Triphlorethol A (5) – White powder; ESI-MS: m/z 373 [M−H]−; 1H and 13C-NMR data, see reference.33
Dieckol (6) – Light brown powder; ESI-MS: m/z 741 [M−H]−; 1H-NMR (DMSO-d6, 400 MHz): δH 9.69 (1H, s, OH-4), 9.59 (1H, s, OH-9′′), 9.49 (1H, s, OH-4′′), 9.44 (1H, s, OH-4), 9.35 (2H, s, OH-3′′′, OH-5′′′), 9.27 (1H, s, OH-2′′), 9.22 (1H, s, OH-2), 9.21 (1H, s, OH-7′′), 9.15 (2H, s, OH-3′, OH-5′), 6.16 (1H, s, H-3′′), 6.14 (1H, s, H-3), 6.02 (1H, d, J = 2.4 Hz, H-8), 5.99 (1H, d, J = 2.4 Hz, H-8′′), 5.95 (2H, s, H-2′′′, H-6′′′), 5.83 (1H, d, J = 2.4 Hz, H-6), 5.82 (1H, d, J = 2.4 Hz, H-6′′), 5.80 (1H, t, J = 2.0 Hz, H-4′), 5.72 (2H, d, J = 2.0 Hz, H-2′, H-6′); 13C-NMR (DMSO-d6, 100 MHz): δC 160.5 (C-1′), 159.0 (C-5′), 156.1 (C-1′′′), 154.4 (C-7), 153.3 (C-7′′), 151.4 (C-3′′′, C-5′′′), 146.3 (C-2), 146.2 (C-9′′), 146.1 (C-9), 146.1 (C-2′′), 142.8 (C-5a′′), 142.6 (C-5a), 142.2 (C-4′′), 142.1 (C-4), 137.4 (C-10a), 137.2 (C-10′′), 124.3 (C-4′′′), 124.1 (C-9a), 123.4 (C-4a), 123.3 (C-4a′′), 122.7 (C-9a′′), 122.4 (C-1′′), 122.3 (C-1), 98.6 (C-8′′), 98.4 (C-3), 98.3 (C-3′′), 98.2 (C-8), 96.3 (C-4′), 94.6 (C-2′′′, C-6′′′), 94.0 (C-6′), 93.7 (C-6).31
2-Phloroeckol (7) – Light brown powder; ESI-MS: m/z 495 [M−H]−; 1H and 13C-NMR data, see reference.32
2-O-(2,4,6-Trihydroxyphenyl)-6,6′-bieckol (8) – Amorphous powder; ESI-MS: m/z 865 [M−H]−; 1H and 13C-NMR data, see reference.32
8,8′-Bieckol (9) – Brown amorphous powder; ESI-MS: m/z 741 [M−H]−; 1H and 13C-NMR data, see reference.32
6,8′-Bieckol (10) – Brown amorphous powder; ESI-MS: m/z 741 [M−H]−; 1H and 13C-NMR data, see reference.32
Inhibitory activity on cholinesterase – The inhibitory activities of ChE were measured using the spectrophotometric method, following the protocol we previously described.34 ACh and BuCh were used as the substrates to assay the inhibitions of AChE and BuChE, respectively. The hydrolysis of ACh or BCh was monitored by following the formation of the yellow 5-thio-2-nitrobenzoate anion at 405 nm, which resulted from the reaction of DTNB with thiocholine, released by the enzymatic hydrolysis of either ACh or BCh, respectively. In a 96-well plate, 130 μL of 0.0325 U/mL AChE in buffer (100 mM potassium phosphate buffer, pH 7.45), 20 μL of sample, 25 μL of 0.125 mM ATCI (acetylthiocholine iodide), 25 μL of 0.625 mM DTNB (5,5-dithiobis-2-nitrobenzoic acid) were added. The absorbance was measured for 1 h at an interval of 30 s at 37oC and 405 nm. The initial reaction rates at the time of the presence of the sample were compared based on the initial reaction rate in the absence of the sample. The results were expressed in percentages. Enzyme inhibition assay for BChE was performed by the same method as that of AChE. Tacrine was used as a positive control,34 and IC50 was applied to samples showing inhibitory effects. The percentage (%) inhibition was calculated as follows:
% inhibition = [(Ac − As) / Ac] × 100, where Ac is the absorbance of the control, and As is the absorbance of the sample.
Molecular docking – Molecular docking simulations were performed to explore the interaction between ligand molecules and the target enzyme using AutoDock 4.2 (The Scripps Research Institute, La Jolla, CA, USA) following our previously described procedure.35-37 The 3D structures of active compounds were generated, and MM2 energy minimization was performed using Spartan′18 software (Wavefunction, CA, USA). The 3D structure of AChE and BuChE were downloaded from RCSB Protein Data Bank with PDB ID: 1C2B and 1P0I, respectively.38,39 Subsequently, hydrogen atoms were incorporated, and Gasteiger charges were assigned using AutoDockTools 1.5.6. The docking settings adhered to the default values, with the exception of Lamarckian genetic algorithms (runs = 100) and a maximum of 25,000,000 evaluations. Ligand interaction diagrams were visualized using BIOVIA Discovery Studio Visualizer (Dassault Systèmes, CA, USA).
Statistical analysis – The results were expressed as means ± SEM for three independently conducted experiments. Statistical significance (p < 0.05) was evaluated using one-way analysis of variance (ANOVA) in conjunction with Duncan′s tests.
Results and Discussion
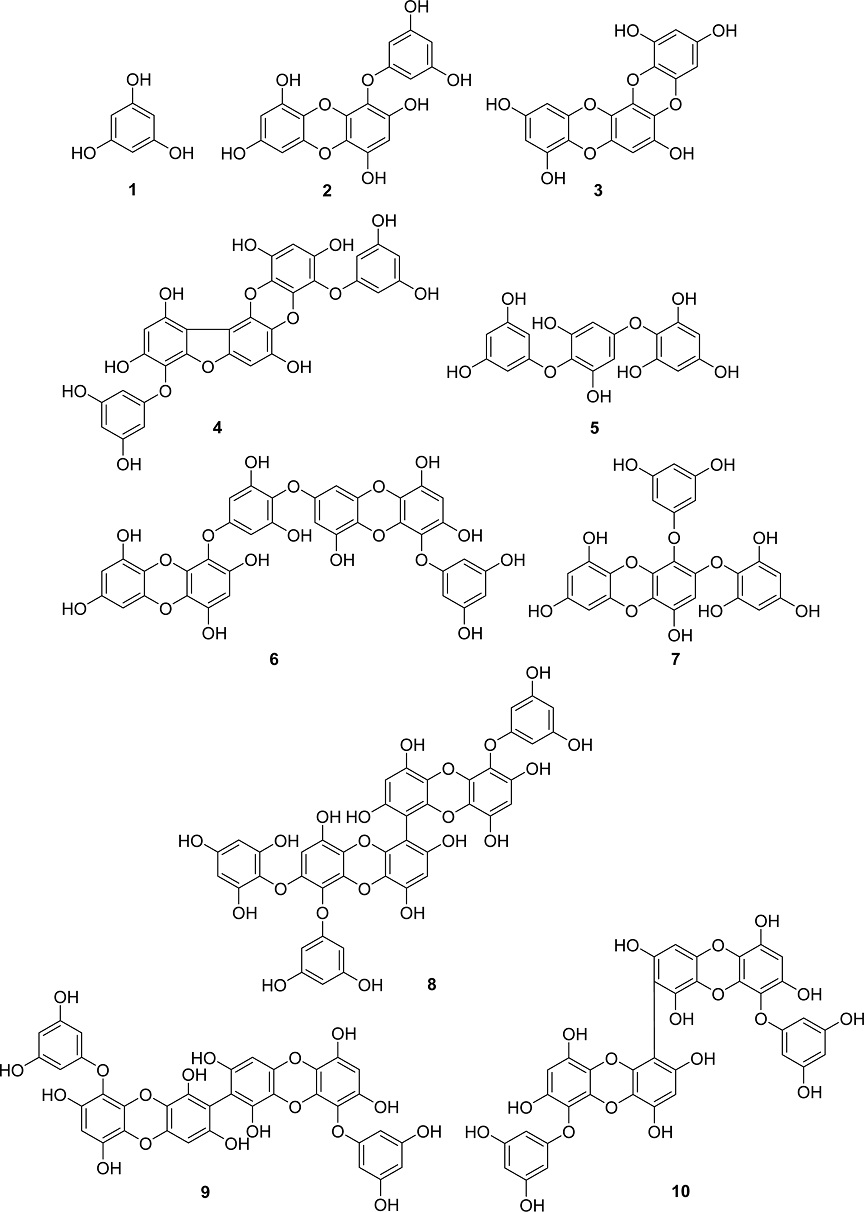
An 80% EtOH extract of E. cava was subject to repeated column chromatography, resulting in the isolation of 10 compounds (1–10). Comparing our spectroscopic findings with published data, the known metabolites were successfully identified as phloroglucinol (1),30 eckol (2),31,32 dioxinodehydroeckol (3),31 phlorofucofuroeckol A (4),31 triphlorethol A (5),33 dieckol (6),31 2-phloroeckol (7),32 2-O-(2,4,6-trihydroxyphenyl)-6,6′-bieckol (8),32 8,8′-bieckol (9),32 and 6,8′-bieckol (10)32 (Fig. 1).
Next, their AChE and BuChE inhibitory activities were evaluated. Tacrine, used as a positive control, had an IC50 value of 118.9 ± 4.6 nM for AChE inhibition and 8.9 ± 2.4 nM for BuChE inhibition. Of the tested compounds, 4, 6, and 9 strongly inhibited AChE, with IC50 values of 0.9 ± 0.8, 4.4 ± 4.7, and 1.4 ± 6.9 µM, respectively. Compounds 1, 2, 7, 8, and 10 displayed moderate AChE inhibition, with IC50 values ranging from 21.6 ± 0.1 to 66.5 ± 0.4 µM (Table 1). Conversely, compounds 3 and 5 showed weak or no inhibition against AChE. In the BuChE inhibition assay, compounds 4, 6, and 9 showed strong inhibitory activity, with IC50 values of 1.4 ± 3.8, 6.4 ± 2.6, and 25.2 ± 0.1 µM, respectively. Compounds 3 and 8 exhibited weak BuChE inhibitory effects, with IC50 values of 92.3 ± 4.7 and 81.0 ± 0.2 µM. The remaining compounds did not inhibit BuChE.
Enzyme kinetics experiments were conducted to determine the type of inhibition and the inhibition constant (Ki value) between the active compounds and target enzyme, using the Lineweaver–Burk and Dixon graphical techniques.40 In the Lineweaver–Burk plot technique, the convergence of inhibitor lines in the xy region signifies mixed inhibition, whereas lines intersecting at the same point on either the x-axis or y-axis denote noncompetitive or competitive inhibition, respectively.41
A previous study discovered that compounds 2, 6, and 9 acted as competitive AChE inhibitors, with Ki values of 37.3, 12.3, and 11.4 µM, respectively.42 In this study, we investigated the AChE inhibition mode of the remaining active compounds from E. cava (4, 7, 8, and 10). Compounds 4, 7, and 8 had Lineweaver–Burk plots consisting of a set of straight lines that intersected with each other in the xy region, indicating that these compounds follow a mixed inhibition mode on AChE (Fig. 2). In comparison, compound 10 had a group of straight lines that intersected each other on the 1/V axis or y-axis, indicating that this metabolite acts as a competitive AChE inhibitor. The inhibition constants of the active AChE compounds were determined from the Dixon plots. The Ki values of compounds 4, 7, 8, and 10 were 4.5, 26.6, 34.6, and 5.4 µM. In the kinetics of BuChE, compounds 4 and 6 acted as mixed BuChE inhibitors, with Ki values of 2.1 and 3.7 µM, respectively (Fig. 3, whereas compound 9 competitively inhibited BuChE, with a calculated Ki value of 31.2 µM.
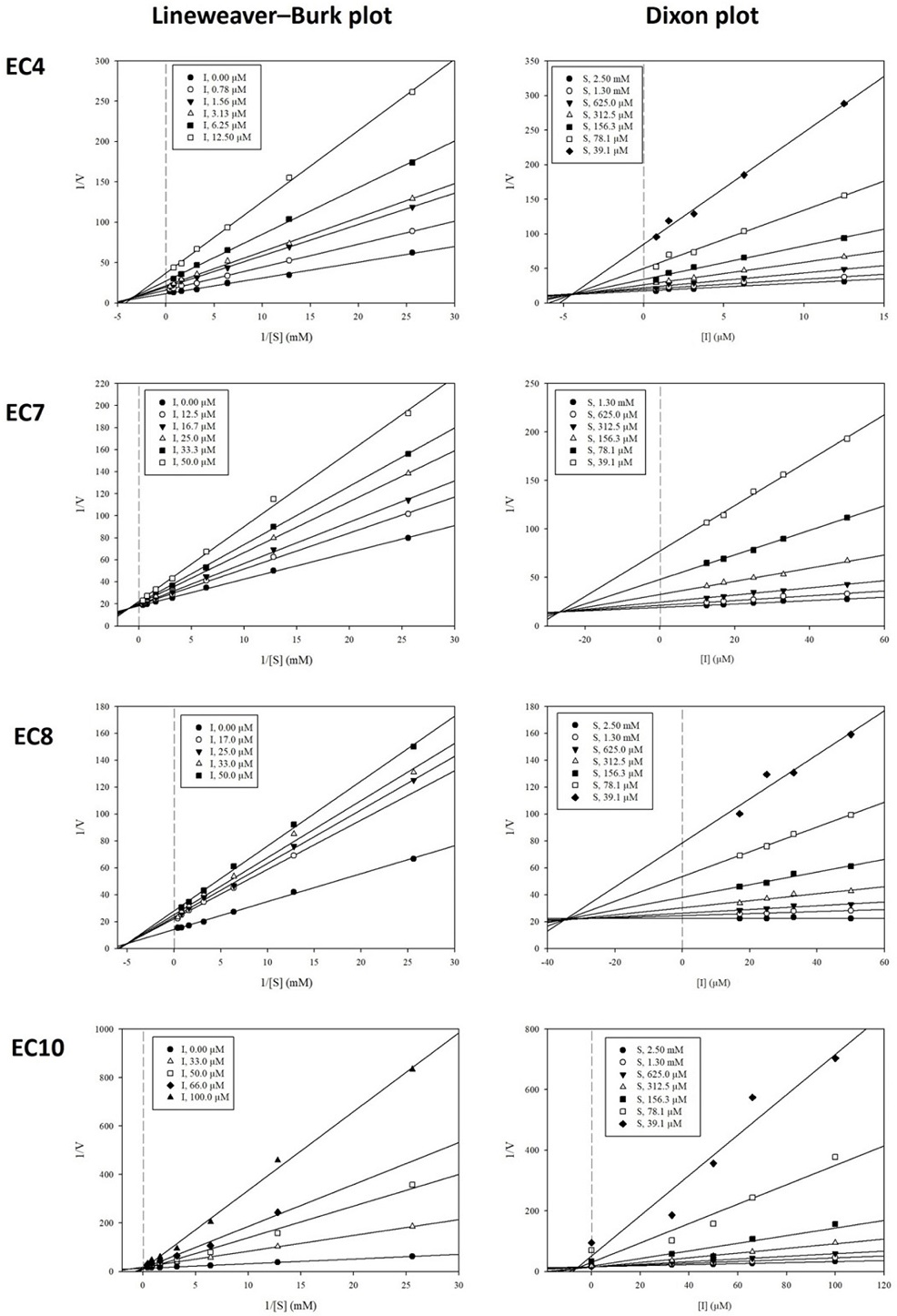
Lineweaver−Burk plots and Dixon plots for the AChE inhibition of active phlorotannins 4, 7, 8, and 10, respectively.
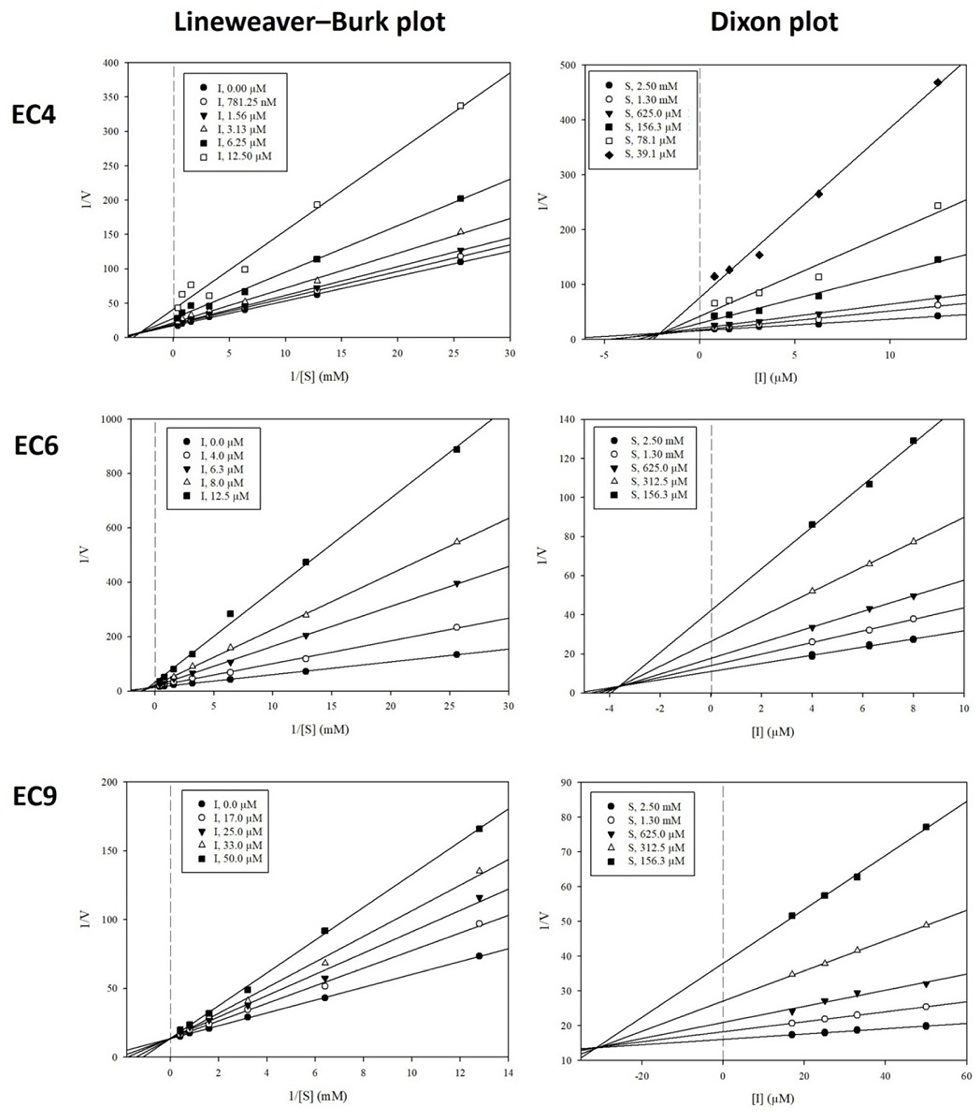
Lineweaver−Burk plots and Dixon plots for the BuChE inhibition of active phlorotannins 4, 6, and 9, respectively.
Molecular docking is widely used for structure-based drug discovery.43-45 In this method, models are used to depict the complicated interactions that occur at the atomic level between a protein and a small molecule. This provides insights into the activity of small molecules at specific binding sites on target proteins, shedding light on essential biochemical processes. Virtual screening is cost-effective and remarkably adaptable to different compounds, which collectively expedite the identification of suitable protein inhibitors.46,47 A molecular docking simulation was used to examine the critical interactions among and binding mechanisms of active compounds with AChE and BuChE inhibitory activity.
The AChE active site is made up of the catalytic triad Pro440-Leu327-Phe200.48,49 A remarkable feature of the AChE structure is its narrow active-site gorge, around 20Å in length, which penetrates the enzyme core. This gorge is defined by a specific set of amino acid residues, including Glu285 and Leu273 in the uppermost area, Tyr72 and Glu334 near the middle, and Leu199 closer to the base.48,50 In comparison, there are three distinct allosteric regions of AChE: the “back door” (Trp86, Gly448, Tyr449, and Ile451), “side door” (Asp74, Thr75, Leu76, Thr83, Glu84, and Asn87), and “acyl loop door” (Trp236, Arg247, and Phe297).51,52 As shown in Fig. 4, the docking simulation revealed that compounds 4, 7, and 8 could bind to the acyl loop door region of AChE. Compound 4 forms hydrogen bonds with Thr238 and Ser240 through its substituent groups. The aromatic rings of this compound displayed π-alkyl interactions with Pro235 and Arg296, π-anion interactions with Glu313 and Arg247, and van der Waals interactions with the crucial amino acid Trp236. Compound 7 established five hydrogen bonds with Thr238, Glu313, Asn533, His405, and Asp404 via methoxy groups. The aromatic rings had π-alkyl interactions with Pro410 and Pro537. Other amino acid residues, including Asn233, Gly234, Pro235, Val239, Thr311, Asn406, Cys409, Trp532, and Leu536, bonded with compound 7 via Van der Waals interactions. Compound 8 formed hydrogen bonds with Gly234, Gln413, and Asn533 via methoxy groups. Its aromatic rings exhibited π-alkyl interactions with Pro537, Pro410, and Arg290, and π-anion interactions with Glu313 and the important residue Arg247 in the allosteric site of AChE. The compound also interacted with Asn233, Thr238, Pro235, Val239, Ser240, Glu243, Leu289, Gln291, Arg296, Thr311, Gln369, Asp404, His405, Cys409, Trp532, Arg534, and Leu540 via van der Waals interactions. As a competitive inhibitor of AChE, compound 10 could bind to the active-site gorge of the target enzyme. Methoxy groups established hydrogen bonds with Ser293, Arg296, Phe338, and Gly342. The aromatic rings of the compound showed π-π stacked interactions with Trp286 and Tyr341, and a π-anion interaction with Glu292. Other amino acids around the active-site gorge of AChE, including Tyr72, Asp74, Leu76, Tyr124, Leu289, Gln291, Ile294, Phe295, Phe297, and Tyr337, formed van der Waals interactions with compound 10. These interactions underscore the stability of compound 10 within the active-site gorge of the target enzyme.
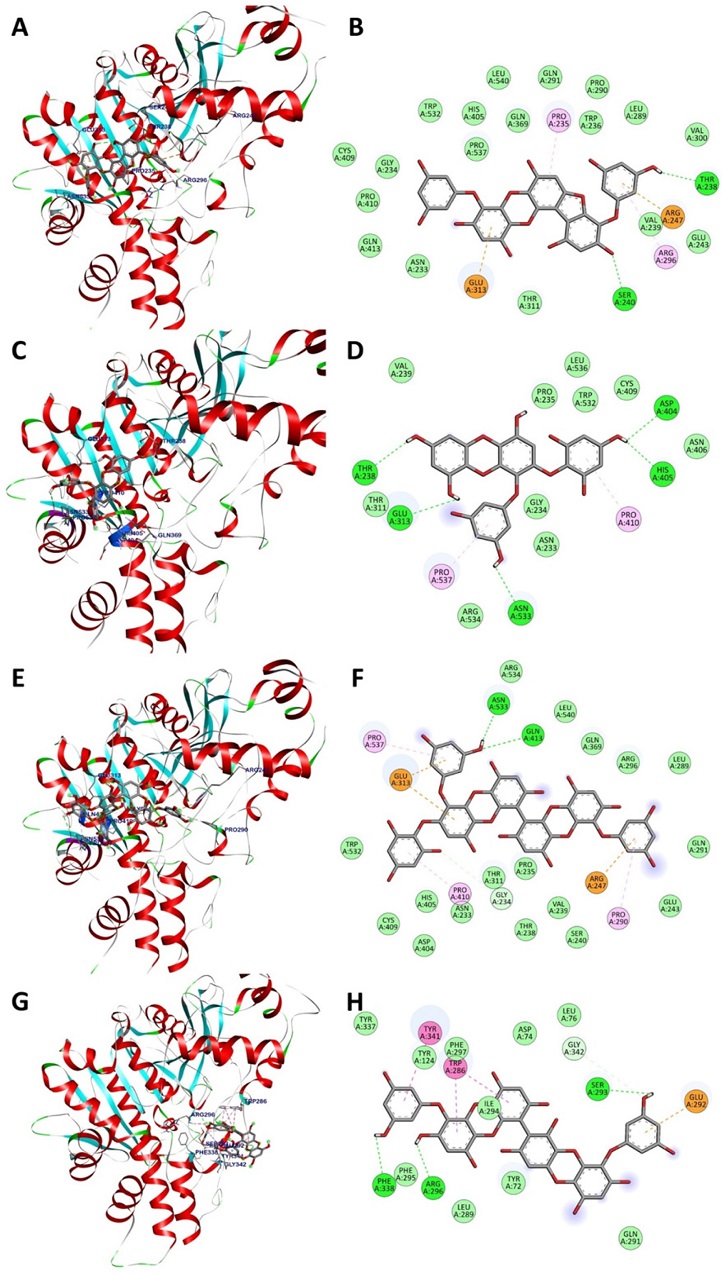
3D docking poses (A, C, E, and G) and interaction diagrams (B, D, F, and H) of AChE inhibition by phlorotannins 4, 7, 8, and 10, respectively. Green: hydrogen bonds, light green: van der Waals interactions, pink: hydrophobic interactions, and orange: electrostatic interactions with the corresponding amino acid residues.
The active site gorge of BuChE includes several key components, namely, the catalytic triad (Ser198, His438, and Glu325), acyl loop (Ala277, Leu286, and Val288), π-cation site (Tyr82 and Ala328), ω-loop (Ile69 to Ser79), oxyanion hole (Ala199, Gly116, and Gly117), and peripheral site (Asn68, Glu70, and Tyr332).45,53 Compounds 4 and 6 showed mixed inhibition of BuChE. Consequently, blind docking, a potent method for investigating receptor binding sites and the corresponding ligand binding poses, was used.54 As presented in Fig. 5, compound 4 could bind at the entrance of the acyl loop, establishing five hydrogen bonding interactions with residues Ala229, Val233, Glu238, Phe357, and Tyr396 and an alkyl interaction with Val280. Other residues, including Phe227, Asn228, Pro230, Trp231, Ala232, Thr234, Ser235, Arg242, Pro281, Tyr282, Gly283, Thr284, Ser287, Met302, Pro303, Asp304, and Pro359, associated with compound 4 through Van der Waals interactions. Where compound 6 displayed close interactions with the acyl loop of BuChE, it established a hydrogen bond with the essential amino acid residue Val288 via its methoxy groups and an oxygen bridge. In addition, van der Waals interactions were observed with the Leu286 residue. Compound 6 also formed hydrogen bonds with Asn228, Tyr396, and Ser287. The presence of aromatic rings promoted hydrophobic interactions with Val280, Pro359, Pro527, and Pro230. In comparison, compound 9 is a competitive inhibitor of BuChE, as revealed by enzyme kinetics assays. This suggests that the compound has the potential to bind to the active site of the target enzyme. The docking simulations showed that this compound could engage with the catalytic triad of BuChE via interactions with residues Ser198 and His438. In addition, it formed essential connections with amino acid residues Asn68 and Asp70 in the peripheral site, as well as with residues Gly116 and Gly117 in the oxyanion hole of BuChE via van der Waals interactions. The compound also linked with the acyl loop of the target enzyme through a π-alkyl interaction with Ala277 and van der Waals interaction with Leu286. Furthermore, it formed hydrogen bonds with Pro285, Asn289, and Ser287, while engaging in alkyl interactions with residue Ala328. Our docking results completely concur with the results of the kinetic studies, confirming the inhibition mode of the active compounds within the binding site of the target enzyme. In addition, molecular docking simulations involving these active compounds, AChE, and BuChE suggest that phlorotannins introduce a new molecular structure, which can be used to explore and advance AChE and BuChE inhibitors.
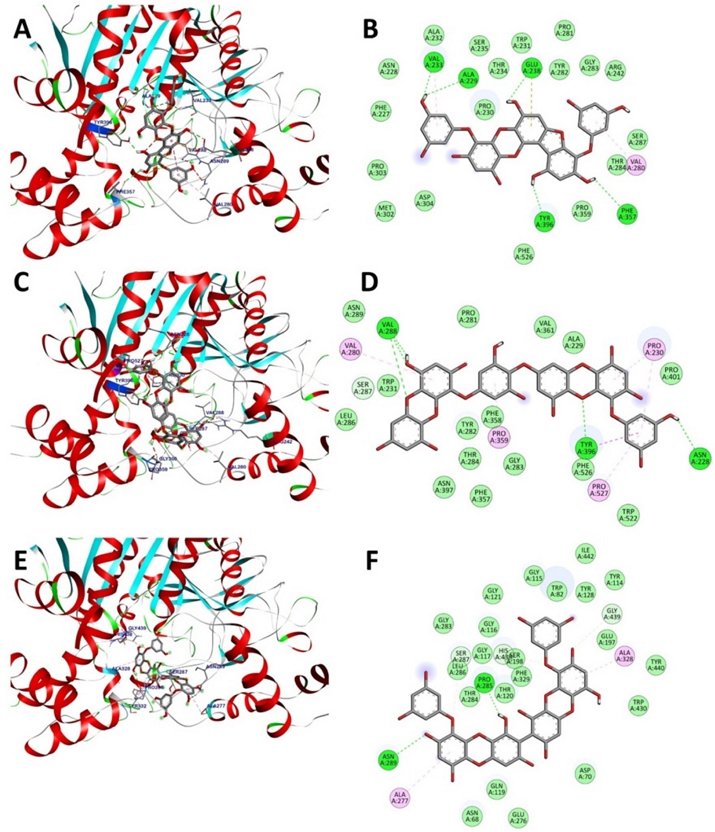
3D docking poses (A, C, and E) and interaction diagrams (B, D, and F) of BuChE inhibition by phlorotannins 4, 6, and 9, respectively.
In summary, a chemical investigation of the marine alga E. cava resulted in the isolation and structural elucidation of 10 phlorotannins (1–10). The inhibitory effects of compounds 1–10 against AChE and BuChE were investigated. Of these, phlorofucofuroeckol A (4) had the most potent inhibition of both AChE and BuChE, with IC50 values of 0.9 ± 0.8 and 1.4 ± 3.8 µM, respectively. Compounds 7, 8, and 10 had moderate inhibitory effects on AChE, whereas compounds 6 and 9 showed inhibition was high of BuChE. Enzyme kinetics analysis was also conducted to investigate the binding inhibition and inhibition constants of the active compounds with the target enzyme. Molecular docking simulations demonstrated that these active compounds bound tightly to the target enzyme. Collectively, our in vitro and in silico findings suggest that phlorotannins derived from E. cava have potential to function as anti-ChE therapeutics against AD.
Acknowledgments
This manuscript is an additional study, and a portion is based on the first author’s master dissertation from Chungnam National University. This study was supported by National Research Foundation of Korea (NRF) funded by the Korea government (MSIT) (No. NRF- 2022R1C1C1004636).
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
-
Breijyeh, Z.; Karaman, R. Molecules 2020, 25, 5789.
[https://doi.org/10.3390/molecules25245789]

-
Andrade-Moraes, C. H.; Oliveira-Pinto, A. V.; Castro-Fonseca, E.; da Silva, C. G.; Guimarães, D. M.; Szczupak, D.; Parente-Bruno, D. R.; Carvalho, L. R. B.; Polichiso, L.; Gomes, B. V.; Oliveira, L. M.; Rodriguez, R. D.; Leite, R. E. P.; Ferretti-Rebustini, R. E. L.; Jacob-Filho, W.; Pasqualucci, C. A.; Grinberg, L. T.; Lent, R. Brain 2013, 136, 3738-3752.
[https://doi.org/10.1093/brain/awt273]

-
Long, J. M.; Holtzman, D. M. Cell 2019, 179, 312-339.
[https://doi.org/10.1016/j.cell.2019.09.001]

-
Ferreira-Vieira, T. H.; Guimaraes, I. M.; Silva, F. R.; Ribeiro, F. M. Curr. Neuropharmacol. 2016, 14, 101-115.
[https://doi.org/10.2174/1570159X13666150716165726]

-
Colovic, M. B.; Krstic, D. Z.; Lazarevic-Pasti, T. D.; Bondzic, A. M.; Vasic, V. M. Curr. Neuropharmacol. 2013, 11, 315-335.
[https://doi.org/10.2174/1570159X11311030006]

-
Grossberg, G. T. Curr. Ther. Res. Clin. Exp. 2003, 64, 216-235.
[https://doi.org/10.1016/S0011-393X(03)00059-6]

-
Thomas, S. J.; Grossberg, G. T. Clin. Interv. Aging 2009, 4, 367-377.
[https://doi.org/10.2147/CIA.S6666]

-
Marucci, G.; Buccioni, M.; Ben, D. D.; Lambertucci, C.; Volpini, R.; Amenta, F. Neuropharmacology 2021, 190, 108352.
[https://doi.org/10.1016/j.neuropharm.2020.108352]

-
Cummings, J. L. Am. J. Psychiatry 2000, 157, 4-15.
[https://doi.org/10.1176/ajp.157.1.4]

-
Mushtaq, G.; Greig, N. H.; Khan, J. A.; Kamal, M. A. CNS Neurol. Disord. Drug Targets 2014, 13, 1432-1439.
[https://doi.org/10.2174/1871527313666141023141545]

-
Greig, N. H.; Lahiri, D. K.; Sambamurti, K. Int. Psychogeriatr. 2002, 14, 77-91.
[https://doi.org/10.1017/S1041610203008676]

-
Ha, M. T.; Park, S. E.; Kim, J. A.; Woo, M. H.; Choi, J. S.; Min, B. S. Nat. Prod. Sci. 2022, 28, 138-142.
[https://doi.org/10.20307/nps.2022.28.3.138]

-
Vecchio, I.; Sorrentino, L.; Paoletti, A.; Marra, R.; Arbitrio, M. J. Cent. Nerv. Syst. Dis. 2021, 13, 11795735211029113.
[https://doi.org/10.1177/11795735211029113]

-
El-Beltagi, H. S.; Mohamed, A. A.; Mohamed, H. I.; Ramadan, K. M. A.; Barqawi, A. A.; Mansour, A. T. Mar. Drugs 2022, 20, 342.
[https://doi.org/10.3390/md20060342]

-
Kang, J.-I.; Kim, S.-C.; Kim, M.-K.; Boo, H.-J.; Jeon, Y.-J.; Koh, Y.-S.; Yoo, E.-S.; Kang, S.-M.; Kang, H.-K. Int. J. Mol. Sci. 2012, 13, 6407-6423.
[https://doi.org/10.3390/ijms13056407]

-
Kang, S.-M.; Heo, S.-J.; Kim, K.-N.; Lee, S.-H.; Jeon, Y.-J. J. Funct. Foods 2012, 4, 158-166.
[https://doi.org/10.1016/j.jff.2011.10.001]

-
Woo, H.; Kim, M. K.; Park, S.; Han, S. H.; Shin, H. C.; Kim, B. G.; Oh, S. H.; Suh, M. W.; Lee, J. H.; Park, M. K. Mar. Drugs 2021, 19, 443.
[https://doi.org/10.3390/md19080443]

-
Park, J. Y.; Kim, J. H.; Kwon, J. M.; Kwon, H. J.; Jeong, H. J.; Kim, Y. M.; Kim, D.; Lee, W. S.; Ryu, Y. B. Bioorg. Med. Chem. 2013, 21, 3730-3737.
[https://doi.org/10.1016/j.bmc.2013.04.026]

-
Maheswari, V.; Babu, P. A. S. Russ. J. Mar. Biol. 2022, 48, 309-324.
[https://doi.org/10.1134/S1063074022050169]

-
Koirala, P.; Jung, H. A.; Choi, J. S. Arch. Pharm. Res. 2017, 40, 981-1005.
[https://doi.org/10.1007/s12272-017-0948-4]

-
Kumar, L. R. G.; Paul, P. T.; Anas, K. K.; Tejpal, C. S.; Chatterjee, N. S.; Anupama, T. K.; Mathew, S.; Ravishankar, C. N. J. Appl. Phycol. 2022, 34, 2173-2185.
[https://doi.org/10.1007/s10811-022-02749-4]

-
Catarino, M. D.; Amarante, S. J.; Mateus, N.; Silva, A. M. S.; Cardoso, S. M. Foods 2021, 10, 1478.
[https://doi.org/10.3390/foods10071478]

-
Besednova, N. N.; Andryukov, B. G.; Zaporozhets, T. S.; Kuznetsova, T. A.; Kryzhanovsky, S. P.; Ermakova, S. P.; Galkina, I. V.; Shchelkanov, M. Y. Mar. Drugs 2022, 20, 243.
[https://doi.org/10.3390/md20040243]

-
Sanjeewa, K. K. A.; Kim, E. A.; Son, K. T.; Jeon, Y. J. J. Photochem. Photobiol. B. 2016, 162, 100-105.
[https://doi.org/10.1016/j.jphotobiol.2016.06.027]

-
Catarino, M. D.; Fernandes, I.; Oliveira, H.; Carrascal, M.; Ferreira, R.; Silva, A. M. S.; Cruz, M. T.; Mateus, N.; Cardoso, S. M. Int. J. Mol. Sci. 2021, 22, 7604.
[https://doi.org/10.3390/ijms22147604]

-
Erpel, F.; Mateos, R.; Pérez-Jiménez, J.; Pérez-Correa, J. R. Food Res. Int. 2020, 137, 109589.
[https://doi.org/10.1016/j.foodres.2020.109589]

-
Li, Y.; Lee, S. H.; Le, Q. T.; Kim, M. M.; Kim, S. K. J. Agric. Food Chem. 2008, 56, 12073-12080.
[https://doi.org/10.1021/jf802732n]

-
Karadeniz, F.; Kang, K. H.; Park, J. W.; Park, S. J.; Kim, S. K. Biosci. Biotechnol. Biochem. 2014, 78, 1151-1158.
[https://doi.org/10.1080/09168451.2014.923282]

-
Langarizadeh, M. A.; Abiri, A.; Ghasemshirazi, S.; Foroutan, N.; Khodadadi, A.; Faghih-Mirzaei, E. Biotechnol. Appl. Biochem. 2021, 68, 918-926.
[https://doi.org/10.1002/bab.2014]

-
Hattori, M.; Shu, Y. Z.; El-Sedawy, A. I.; Namba, T.; Kobashi, K.; Tomimori, T. J. Nat. Prod. 1988, 51, 874-878.
[https://doi.org/10.1021/np50059a010]

-
Kang, H. S.; Chung, H. Y.; Jung, J. H.; Son, B. W.; Choi, J. S. Chem. Pharm. Bull. 2003, 51, 1012-1014.
[https://doi.org/10.1248/cpb.51.1012]

-
Park, S. R.; Kim, J. H.; Jang, H. D.; Yang, S. Y.; Kim, Y. H. Food Chem. 2018, 257, 128-134.
[https://doi.org/10.1016/j.foodchem.2018.03.013]

-
Yoon, M.; Cho, S. Mar. Drugs 2018, 16, 139.
[https://doi.org/10.3390/md16050139]

-
Vinh, L. B.; Dan, G.; Phong, N. V.; Cho, K.; Kim, Y. H.; Yang, S. Y. Chem. Nat. Compd. 2021, 57, 784-787.
[https://doi.org/10.1007/s10600-021-03478-9]

-
Phong, N. V.; Yang, S. Y.; Min, B. S.; Kim, J. A. J. Mol. Struct. 2023, 1282, 135188.
[https://doi.org/10.1016/j.molstruc.2023.135188]

-
Manh Khoa, N.; Viet Phong, N.; Yang, S. Y.; Min, B. S.; Kim, J. A. Bioorg. Chem. 2023, 134, 106464.
[https://doi.org/10.1016/j.bioorg.2023.106464]

-
Phong, N. V.; Zhao, Y.; Min, B. S.; Yang, S. Y.; Kim, J. A. Metabolites 2022, 12, 938.
[https://doi.org/10.3390/metabo12100938]

-
Bourne, Y.; Grassi, J.; Bougis, P. E.; Marchot, P. J. Biol. Chem. 1999, 274, 30370-30376.
[https://doi.org/10.1074/jbc.274.43.30370]

-
Nicolet, Y.; Lockridge, O.; Masson, P.; Fontecilla-Camps, J. C.; Nachon, F. J. Biol. Chem. 2003, 278, 41141-41147.
[https://doi.org/10.1074/jbc.M210241200]

-
Wang, J.; Araki, T.; Ogawa, T.; Matsuoka, M.; Fukuda, H. Biotechnol. Bioeng. 1999, 62, 402-411.
[https://doi.org/10.1002/(SICI)1097-0290(19990220)62:4<402::AID-BIT3>3.0.CO;2-V]

-
Phong, N. V.; Oanh, V. T.; Yang, S. Y.; Choi, J. S.; Min, B. S.; Kim, J. A. Int. J. Biol. Macromol. 2021, 188, 719-728.
[https://doi.org/10.1016/j.ijbiomac.2021.08.091]

-
Lee, J.; Jun, M. Mar. Drugs 2019, 17, 91.
[https://doi.org/10.3390/md17020091]

-
Trang, N. M.; Kim, E. N.; Pham, T. H.; Jeong, G. S. J. Agric. Food Chem. 2023, 71, 10037-10049.
[https://doi.org/10.1021/acs.jafc.3c00368]

-
Maurya, R.; Boini, T.; Misro, L.; Radhakrishnan, T. Nat. Prod. Sci. 2023, 29, 104-112.
[https://doi.org/10.20307/nps.2023.29.2.104]

-
Wu, Y.; Su, X.; Lu, J.; Wu, M.; Yang, S. Y.; Mai, Y.; Deng, W.; Xue, Y. Front. Pharmacol. 2022, 13, 905708.
[https://doi.org/10.3389/fphar.2022.905708]

-
Trang, N. M.; Kim, E. N.; Lee, H. S.; Jeong, G. S. Biomolecules 2022, 12, 256.
[https://doi.org/10.3390/biom12020256]

-
Sadybekov, A. V.; Katritch, V. Nature 2023, 616, 673-685.
[https://doi.org/10.1038/s41586-023-05905-z]

-
Dvir, H.; Silman, I.; Harel, M.; Rosenberry, T. L.; Sussman, J. L. Chem. Biol. Interact. 2010, 187, 10-22.
[https://doi.org/10.1016/j.cbi.2010.01.042]

-
Xu, Y.; Cheng, S.; Sussman, J. L.; Silman, I.; Jiang, H. Molecules 2017, 22, 1324.
[https://doi.org/10.3390/molecules22081324]

-
Xu, Y.; Colletier, J. P.; Weik, M.; Jiang, H.; Moult, J.; Silman, I.; Sussman, J. L. Biophys. J. 2008, 95, 2500-2511.
[https://doi.org/10.1529/biophysj.108.129601]

-
Bennion, B. J.; Essiz, S. G.; Lau, E. Y.; Fattebert, J. L.; Emigh, A.; Lightstone, F. C. PLoS One 2015, 10, e0121092.
[https://doi.org/10.1371/journal.pone.0121092]

-
Roca, C.; Requena, C.; Sebastián-Pérez, V.; Malhotra, S.; Radoux, C.; Pérez, C.; Martinez, A.; Antonio Páez, J.; Blundell, T. L.; Campillo, N. E. J. Enzyme Inhib. Med. Chem. 2018, 33, 1034-1047.
[https://doi.org/10.1080/14756366.2018.1476502]

-
Vyas, S.; Beck, J. M.; Xia, S.; Zhang, J.; Hadad, C. M. Chem. Biol. Interact. 2010, 187, 241-245.
[https://doi.org/10.1016/j.cbi.2010.04.004]

-
Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z. X.; Cao, Y. Nucleic Acids Res. 2022, 50, W159-W164.
[https://doi.org/10.1093/nar/gkac394]