Inhibitory Activity of PTP1B and α-Glucosidase by Compounds from Whole Plants of Houttuynia cordata Thunb.
Abstract
In this study, twenty known compounds were isolated from Houttuynia cordata Thunb., including four megastigmanes (1‒4), four phenolics (5, 6, 9, and 10), one tetrahydro-2-one derivative (12), four coumarins (7, 13, 14, and 16), six caffeic acid derivatives (8, 11, 15, 17, 18, and 19), and one triterpenoid (20). Their chemical structures were established using NMR spectra and comparison with literature. The anti-diabetic activity of the isolated compounds was assessed by investigating their inhibitory effects on protein tyrosine phosphatase 1B (PTP1B) and α-glucosidase. The results revealed that ginnalin A (10) and 3-(4′-hydroxyphenyl)-2-propenoic acid (4′′-carboxyl)-phenyl ester (13) exhibited significant inhibitory effects on both PTP1B and α-glucosidase with IC50 values of 7.9 ‒ 37.6 and 13.9 ‒ 31.9 μM, respectively. In the kinetic study, these two compounds showed noncompetitive-type PTP1B and α-glucosidase inhibition, with Ki values of 35.6 and 7.3 μM for PTP1B and 13.9 and 31.0 μM for α-glucosidase, respectively. These findings highlight the potential of the isolated compounds as candidates for the development of novel therapeutic agents for diabetes.
Keywords:
Houttuynia cordata, Saururaceae, PTP1B, α-glucosidaseIntroduction
Diabetes mellitus is one of the prioritized non-communicable diseases, along with chronic lung disease, cardiovascular disease, and cancer. The worldwide prevalence of diabetes is projected to increase from 10.5% to 12.2% among individuals aged 20‒79 between 2021 and 2045. By 2045, it is estimated that there will be approximately 783.2 million people with diabetes, compared to 536.6 million in 2021. The highest incidence of diabetes occurs in the age group of 75‒79, with no significant differences between genders.1 In 2021, urban areas have shown a higher prevalence of diabetes cases (12.1%) compared to rural areas (8.3%). This pattern is also observed in high-income countries (11.1%) compared to low-income countries (5.5%). The global health expenditure related to diabetes was approximately $966 billion in 2021, and it is expected to reach $1,054 billion by 2045.2 Tragically, the number of deaths attributed to diabetes has significantly increased over the years. In 2015, 1.5 million individuals lost their lives due to diabetes, while the number rose to 6.7 million in 2021. These statistics highlight the urgent need for effective prevention, management, and treatment strategies to mitigate the burden of diabetes and its associated complications.3 Type 2 diabetes is linked to physical inactivity, obesity, and being overweight, representing over 90% of global diabetes cases. Improper insulin utilization causes elevated blood sugar levels, stimulating insulin release from pancreatic β cells. PTP1B (protein tyrosine phosphatase 1B) enzyme negatively regulates insulin signaling by dephosphorylating insulin substrate and its receptor, affecting insulin function.4 Significantly, bioactive compounds that concurrently inhibit α-glucosidase and PTP1B enzymes display a synergistic effect, effectively reducing hyperglycemia and enhancing insulin sensitization.5
Houttuynia cordata Thunb., a perennial flowering plant belonging to the Houttuynia genus (Saururaceae family), thrives in its native regions of Korea, China, Japan, and Southeast Asia.6 Traditionally, H. cordata holds esteemed value, being highly regarded for its diverse therapeutic applications. It has proven to be effective in treating infectious diseases, malignant pleural effusion, refractory hemoptysis, and nephrotic syndrome. Additionally, its efficacy extends to bronchitis, cough, dropsy, pneumonia, uteritis, eczema, dysentery, and leukorrhea.7,8 Furthermore, H. cordata finds versatile applications in functional food and cosmetic formulations, providing effective solutions to combat various skin issues such as wrinkles, cracked skin, aging signs, acne, melasma, freckles, and skin whitening.6 Bauer et al. have identified essential oils, flavonoids, and alkaloids as prominent compounds in this plant.9 The essential oils have shown remarkable anti-inflammatory, antibacterial, and antiviral properties.10,11 Flavonoids isolated from H. cordata exhibit a diverse range of beneficial effects, including anticancer, antioxidant, anti-mutagenic, and free radical scavenging activities.6,12,13 Furthermore, the alkaloids in H. cordata display potent antiplatelet and cytotoxic activities.14 The primary objective of this study is to investigate bioactive compounds derived from the H. cordata plant that exhibit potential for diabetes treatment. This investigation is grounded in the evaluation of their capacity to inhibit PTP1B and α-glucosidase activity, thereby contributing to the advancement of therapeutic approaches for diabetes management.
Experimental
General experimental procedures ‒ Optical rotations were measured using a JASCO P-2000 digital polarimeter (JASCO Corporation, Tokyo, Japan). UV and electronic circular dichroism (ECD) spectra were obtained using a J-1500 circular dichroism spectrophotometer (JASCO Corporation, Tokyo, Japan). NMR data was acquired using Varian Unity Inova 400 MHz (400 MHz for 1H NMR, 100 MHz for 13C NMR) (Varian, Inc., California, USA) and Bruker Ascend™ 500 MHz (500 MHz for 1H NMR, 125 MHz for 13C NMR) (Bruker Corporation, Massachusetts, USA) spectrometers. For preparative high-performance liquid chromatography (HPLC), a YMC Pak ODS-A column (20 × 250 mm, 5 μM particle size; YMC Co., Ltd., Japan) and a Waters 2487 controller system with a UV detector (UV/VIS - 156) were utilized.
Chemicals and reagents ‒ For open tubular column chromatography, reverse phase RP-18 (Merck, Damstart, Germany, 75 mesh) and normal phase silica gel (Merck, Damstart, Germany, 230–400 mesh) were utilized. Thin-layer chromatography (TLC) was performed using Merck precoated silica gel F254 and RP-18 F254 plates. Solvents of special and first-grade quality were used without requiring purification. High-performance liquid chromatography (HPLC) solvents were obtained from Burdick & Jackson (USA).
Plant materials ‒ In September 2021, the whole plants of H. cordata were collected from the Medicinal Plants Garden of Daegu Catholic University, Korea. The plant was identified by Professor Byung Sun Min, and a voucher specimen (CDU-1006) was deposited at the Herbarium of the College of Pharmacy, Daegu Catholic University, Korea.
Extraction and isolation ‒ The dried whole plants of H. cordata (4.2 kg) were crushed and subjected to methanol extraction (3 times × 10 L) under reflux for 4 hours. This process resulted in a crude extract, which was then filtered and thoroughly dried, yielding 600 g of residue. The obtained residue was dissolved in water (0.5L) and subsequently partitioned with different solvents, including n-hexane (0.5L × 2), dichloromethane (0.5L × 2), ethyl acetate (0.5L × 2), and n-butanol (0.5L × 2). This partitioning procedure successfully produced distinct fractions, namely n-hexane (170 g), dichloromethane (9 g), ethyl acetate (50 g), n-butanol (45g) fractions, and a water layer. The ethyl acetate fraction was subjected to further separation using a silica gel column (5cm × 30 cm) with a stepwise gradient elution of dichloromethane-ethyl acetate (30:1 → 0:1, each 2 L). This resulted in the formation of four fractions, namely E1 to E4. In detail, fraction E1 (4.00 g) was purified on a silica gel column (3 cm × 40 cm) using a dichloromethane-ethyl acetate (20:1) solvent system, yielding three sub-fractions, E1A, E1B, and E1C. Sub-fraction E1A (2.00 g) underwent further chromatography on an RP-18 silica gel column (2.5cm × 45 cm) with an acetonitrile-water (7:1) eluent, resulting in the isolation of compound 1 (15 mg) along with three additional fractions, E1A1, E1A2, and E1A3. Further chromatography of sub-fraction E1A1 (0.45g) on a silica gel column (1.5cm × 40 cm) with a dichloromethane-methanol (9:1) mobile phase led to the isolation of compound 2 (20 mg) and three additional fractions, E1A1A, E1A1B, and E1A1C. Further chromatography of fraction E1A1B (0.08 g) was carried out using an HPLC preparation system with a methanol-water (40:60) eluent, resulting in the isolation of compounds 3 (23 mg, tR = 30 min) and 4 (15 mg, tR = 34 min). Additionally, fraction E1A2 (0.80 g) was purified on a silica gel column (2 cm × 45cm) using a dichloromethane-ethyl acetate (8:1) eluent, yielding compound 5 (24 mg) and two fractions, E1A2A and E1A2B. Finally, sub-fraction E1A2A (0.200 g) underwent separation using an HPLC preparation system with a methanol-water (40:60) eluent, resulting in the isolation of compound 6 (11 mg, , tR = 40 min). E1B was purified on a silica gel column (2 cm × 40 cm) using dichloromethane-ethyl acetate (7:1) leading to the isolation of compound 7 (30 mg). Moving on to fraction E2 (5.00 g), fractionation on a silica gel column (4 cm × 35 cm) with a dichloromethane-methanol (10:1) eluent produced four fractions labeled as E2A to E2D. Sub-fraction E2B (0.75g) was further subjected to chromatography on an RP-18 silica gel column (2 cm × 45cm) with a methanol-water (2.5:1) mixture as the eluent, leading to the isolation of compound 8 (24 mg). Fraction E2C (3.00 g) was purified on a silica gel column (2 cm × 40 cm) using a dichloromethane-ethyl acetate (10:1) solvent system, generating three fractions denoted as E2C1, E2C2, and E2C3. Further chromatography of sub-fraction E2C1 (1.00 g) on a silica gel column (2.5cm × 45cm) with a dichloromethane-ethyl acetate (4:1) eluent resulted in the isolation of compound 9 (20 mg) along with three additional fractions, E2C1A, E2C1B, and E2C1C. Subfraction E2C1C (0.25 g) underwent fractionation using an HPLC preparation system with a methanol-water (40:60) mobile phase, yielding compounds 10 (12 mg, tR = 43 min) and 11 (24 mg, tR = 45min). Furthermore, subfraction E2C2 (0.10 g) was subjected to chromatography using an HPLC preparation system with a methanol-water (40:60) eluent, leading to the isolation of compound 12 (20 mg, tR = 37 min). After fractionation, 1.00 g of fraction E2D was purified on an RP-18 silica gel column (2 cm × 45cm) using a methanol-water (1:2) eluent, resulting in the isolation of compound 13 (20 mg) along with three fractions designated as E2D1, E2D2, and E2D3. Further separation of 0.15g of fraction E2D2 was carried out using an HPLC preparation system with a methanol-water (30:70) mobile phase, leading to the isolation of compound 14 (24 mg, tR = 40 min). Additionally, 0.20 g of fraction E2D3 was fractionated using an HPLC preparation system with a methanol-water (30:70) eluent, yielding compound 15 (23 mg, tR = 35min). In addition, 7.00 g of fraction E4 underwent chromatography on a silica gel column (4 cm × 45cm) using an ethyl acetate-methanol (10:1) mixture as the eluent, resulting in the formation of three fractions designated as E4A to E4C. Subfraction E4B (0.35g) was further fractionated using an HPLC preparation system with a methanol-water (30:70) mobile phase, leading to the isolation of compounds 16 (14 mg, tR = 39 min) and 17 (21 mg, tR = 47 min). Furthermore, subfraction E4C (0.25g) was fractionated using an HPLC preparation system with a methanol-water (30:70) eluent, resulting in the formation of compounds 18 (20 mg, tR = 32 min), 19 (19 mg, tR = 42 min), and 20 (15mg, tR = 49 min), respectively.
Staphylionoside H (1) ‒ White amorphous powder; 1H NMR (CD3OD, 400 MHz): δH 6.06 (1H, dd, J = 16.0, 1.0 Hz, H-7), 5.57 (1H, dd, J = 16.0, 6.0 Hz, H-8), 4.51 (1H, qdd, J = 6.0, 6.0, 1.0 Hz, H-9), 4.27 (1H, d, J = 8.0 Hz, H-1′), 3.87 (1H, dd, J = 12.0, 2.0 Hz, Hb-6′), 3.74 (1H, m, H-3), 3.64 (1H, dd, J = 12.0, 6.0 Hz, Ha-6′), 2.32 (1H, dd, J = 14.0, 9.0 Hz, Hb-4), 1.61 (1H, dd, J = 14.0, 4.1 Hz, Ha-4), 1.57 (1H, ddd, J = 13.0, 5.0, 2.0 Hz, Ha-2), 1.25 (3H, s, H-13), 1.27 (3H, d, J = 6.0 Hz, H-10), 1.21 (1H, dd, J = 13.0, 10.0 Hz, Hb-2), 1.13 (3H, s, H-11), 0.95 (3H, s, H3- 12); 13C NMR (CD3OD, 100 MHz): δC 136.2 (C-8), 129.7 (C-7), 101.2 (C-1′), 78.2 (C-3′), 78.1 (C-5′), 75.1 (C-2′), 74.8 (C-9), 71.9 (C-4′), 71.2 (C-6), 68.2 (C-5), 64.7 (C-3), 62.8 (C-6′), 47.7 (C-2), 41.5 (C-4), 35.9 (C-1), 30.2 (C-11), 25.1 (C-12), 22.6 (C-10), 20.6 (C-13).15
Phaseic acid (2) ‒ White amorphous powder; 1H NMR (CD3OD, 400 MHz): δH 8.11 (1H, d, J = 15.9 Hz, H-8), 6.47 (1H, d, J = 15.9 Hz, H-7), 5.77 (1H, brs, H-11), 3.95 (1H, dd, J = 7.6, 2.8 Hz, Ha-15), 3.67 (1H, d, J = 7.6 Hz, Hb-15), 2.81 (1H, d, J = 17.9 Hz, Ha-2), 2.72 (1H, dd, J = 18.0, 2.8 Hz, Ha-4), 2.48 (1H, dd, J = 17.9, 2.4 Hz, Hb-2), 2.37 (1H, dd, J = 18.0, 2.4 Hz, Hb-4), 2.06 (3H, s, H-10), 1.22 (3H, s, H-13), 1.02 (3H, s, H-14); 13C NMR (CD3OD, 100 MHz): δC 210.9 (C-3), 169.6 (C-12), 151.1 (C-9), 133.7 (C-8), 132.8 (C-7), 119.9 (C-11), 87.9 (C-5), 83.1 (C-6), 78.7 (C-15), 54.1 (C-4), 53.3 (C-2), 49.7 (C-1), 21.3 (C-10), 19.5 (C-13), 15.9 (C-14).16
Dehydrovomifoilol (3) ‒ Colorless oil; 1H NMR (CD3OD, 400 MHz): δH 7.02 (1H, d, J = 15.8 Hz, H-7), 6.45 (1H, d, J = 15.8 Hz, H-8), 5.95 (1H, s, H-4), 2.61 (1H, d, J = 17.1 Hz, Ha-2), 2.31 (3H, s, H-10), 2.29 (1H, d, J =17.1 Hz, Hb-2), 1.91 (3H, s, H-13), 1.07 (3H, s, H-11), 1.05 (3H, s, H-12); 13C NMR (CD3OD, 100 MHz): δC 200.7 (C-3), 200.3 (C-9), 164.6 (C-5), 148.3 (C-7), 131.7 (C-8), 128.0 (C-4), 79.9 (C-6), 50.2 (C-2), 42.6 (C-1), 27.6 (C-10), 24.7 (C-11), 23.5 (C-12), 19.1 (C-13).17
Dehydrovomifoilol glycoside (4) ‒ White amorphous powder; 1H NMR (CD3OD, 400MHz): δH 5.95 (1H, br. s, H-4), 5.82 (1H, dd, J =16.0, 5.6 Hz, H-8), 5.76 (1H, d, J =16.0 Hz, H-7), 4.57 (1H, d, J =8.0 Hz, H-l′-glc), 4.48 (1H, m, H-9), 3.92 (1H, d, J = 11.7 Hz, Ha-6′-glc), 3.70 (1H, dd, J = 11.7, 5.13 Hz, Hb -6′-glc), 3.30-3.43 (3H, m, H-3′-glc, H-4′-glc, H-5′-glc), 3.23 (1H, t, J = 8.2 Hz, H-2′-glc), 2.57 (1H, d, J = 17.0 Hz, Hb-2), 2.20 (1H, d, J = 17.0 Hz, Ha-2), 1.97 (3H, br. s, C-13), 1.34 (3H, d, J = 6.41 Hz, H-10), 1.08 (6H, s, C-11, C-12); l3C NMR (CD3OD, 100 MHz): δC 201.21 (C-3), 167.23 (C-5), 135.20 (C-8), 131.50 (C-7), 127.12 (C-4), 102.64 (C-l′-glc), 79.96 (C-6), 77.91 (C-5′-glc), 77.91 (C-3′-glc), 77.27 (C-9), 75.15 (C-4′-glc), 71.54 (C-2′-glc), 62.74 (C-6′-glc), 50.62 (C-2), 42.37 (C-1), 24.66 (C-11), 23.42 (C-10), 21.20 (C-12), 19.56 (C-13).18
Icariside E5 (5) ‒ White amorphous powder; 1H NMR (CD3OD, 400 MHz): δH 6.98 (1H, d, J = 1.6 Hz, H-2′), 6.95 (1H, d, J = 1.6 Hz, H-6′), 6.62 (1H, overlapped, H-5), 6.61 (1H, overlapped, H-7′), 6.58 (1H, overlapped, H-2), 6.52 (1H, dd, J = 8.0, 1.6 Hz, H-6), 6.34 (1H, dt, J = 16.0, 5.8 Hz, H-8′), 4.71 (1H, d, J = 7.3 Hz, H-1′′), 4.26 (2H, d, J = 5.8 Hz, H-9′), 4.01 (1H, m, H-8), 3.87 (3H, s, 3′-OCH3), 3.82 (1H, dd, J = 11.8, 2.3 Hz, Ha-6′′), 3.78 (1H, overlapped, 8-CH2OH), 3.73 (1H, s, 3-OCH3), 3.71 (1H, dd, J = 11.8, 5.2 Hz, Hb-6′′), 3.68 (1H, overlapped, 8-CH2OH), 3.48 (1H, t, J = 8.4 Hz, H-2′′), 3.41 (1H, t, J = 8.4 Hz, H-3′′), 3.42 (1H, t, J = 8.4 Hz, H-4′′), 3.15 (1H, m, H-5′′), 3.01 (1H, dd, J = 13.5, 5.9 Hz, Ha-7), 2.76 (1H, dd, J = 13.5, 9.3 Hz, Hb-7); 13C NMR (CD3OD, 100 MHz): δC 153.5 (C-3′), 148.5 (C-3), 145.6 (C-4), 145.1 (C-4′), 139.1 (C-5′), 135.5 (C-1′), 133.3 (C-1), 131.6 (C-7′), 129.7 (C-8′), 122.9 (C-6), 118.8 (C-6′), 116.1 (C-2), 114.2 (C-5), 108.8 (C-2′), 106.0 (C-1′′), 77.7 (C-5′′), 77.4 (C-3′′), 75.6 (C-2′′), 71.1 (C-4′′), 66.6 (8-CH2OH), 63.6 (C-9′), 62.2 (C-6′′), 56.1 (3-OCH3), 55.8 (3′-OCH3), 43.1 (C-8), 39.2 (C-7).19
Saikolignanoside A (6) ‒ White amorphous powder; 1H NMR (CD3OD, 400 MHz): δH 6.97 (2H, s, H-6′, H-5), 6.60 (1H, overlapped, H-7′), 6.59 (1H, overlapped, H-2), 6.51 (1H, dd, J = 8.0, 1.6 Hz, H-6), 6.33 (1H, dt, J = 16.0, 5.8 Hz, H-8′), 4.72 (1H, d, J = 7.3 Hz, H-1′′), 4.25 (2H, d, J = 5.8 Hz, H-2, H-9′), 4.00 (1H, m, H-8), 3.86 (3H, s, 3′-OCH3), 3.81 (1H, dd, J = 11.8, 2.3 Hz, Ha-6′′), 3.79 (1H, overlapped, 8-CH2OH), 3.72 (1H, s, 3-OCH3), 3.70 (1H, dd, J = 11.8, 5.1 Hz, Hb-6′′), 3.69 (1H, overlapped, 8-CH2OH), 3.47 (1H, t, J = 8.4 Hz, H-2′′), 3.43 (1H, t, J = 8.4 Hz, H-3′′), 3.41 (1H, t, J = 8.4 Hz, H-4′′), 3.14 (1H, m, H-5′′), 3.00 (1H, dd, J = 13.5, 5.9 Hz, Ha-7), 2.75 (1H, dd, J = 13.5, 9.3 Hz, Hb-7); 13C NMR (CD3OD, 100 MHz): δC 153.4 (C-3′), 148.3 (C-3), 145.3 (C-4), 145.0 (C-4′), 139.0 (C-5′), 135.4 (C-1′), 133.2 (C-1), 131.5 (C-7′), 129.6 (C-8′), 122.8 (C-6), 118.7 (C-6′), 116.0 (C-2), 114.1 (C-5), 108.7 (C-2′), 106.0 (C-1′′), 77.8 (C-5′′), 77.5 (C-3′′), 75.7 (C-2′′), 71.0 (C-4′′), 66.5 (8-CH2OH), 63.5 (C-9′), 62.3 (C-6′′), 56.0 (3-OCH3), 55.9 (3′-OCH3), 43.0 (C-8), 39.1 (C-7).20
Salidroside (7) ‒ White amorphous powder; 1H NMR (DMSO-d6, 400 MHz): δH 9.16 (s, 1H, 4-OH), 7.02 (2H, d, J = 8.6 Hz, H-2, H-6), 6.64 (2H, d, J = 8.6 Hz, H-3, H-5), 4.97 (1H, d, J = 5.1 Hz, 6′-OH), 4.92 (1H, d, J = 5.1 Hz, 4′-OH), 4.88 (1H, d, J = 5.1 Hz, 3′-OH), 4.48 (1H, t, J = 5.9 Hz, 2′-OH), 4.14 (1H, d, J = 7.8 Hz, 1′), 3.85 (1H, dt, J = 9.0, 7.0 Hz, Ha-8), 3.64 (1H, ddd, J = 1.8, 11.7, 6.1 Hz, Ha-6′), 3.54 (1H, dt, J = 9.1, 6.7 Hz, Hb-8), 3.40 (1H, dd, J = 11.7, 5.9 Hz, Hb-6′), 3.09 (2H, m, H-3′), 3.05 (1H, m, H-5′), 3.02 (1H, m, H4′), 2.92 (ddd, J = 4.7, 7.8, 8.6 Hz, H-2′), 2.71 (2H, m, H-7); 13C NMR (DMSO-d6, 100 MHz): δC 155.9 (C-4), 130.1 (C-2, C-6), 128.9 (C-1), 115.3 (C-3, C-5), 103.1 (C-1′), 77.2 (C-2′), 77.1 (C-3′), 73.7 (C-5′), 70.4 (C-4′), 70.2 (C-8), 61.4 (C-6′), 35.14 (C-7).21
3,4-dihydroxyphenylethyl alcohol 8-O-β-D-glucopyranoside (8) ‒ Pale yellowish powder; 1H NMR (CD3OD, 400 MHz): δH 6.73 (1H, d, J = 2.1 Hz, H-2), 6.72 (1H, d, J = 8.4 Hz, H-5), 6.64 (1H, dd, J = 8.4, 2.1 Hz, H-6), 4.33 (1H, d, J = 7.7 Hz, H-1′), 4.07 (2H, t, J = 7.3 Hz, H-8), 3.71 (1H, dd, J = 14.7, 7.3 Hz, Ha-7), 3.62 (1H, dd, J = 11.7, 6.1 Hz, Ha-6′), 3.40 (1H, dd, J = 11.7, 6.1 Hz, Hb-6′), 3.28 (2H, m, H-3′, H-5′), 3.25 (1H, m, H-4′), 3.18 (1H, m H-2′), 2.81 (1H, dd, J = 14.7, 7.3 Hz, Hb-7); 13C NMR (CD3OD, 100 MHz): δC 146.5 (C-3), 145.0 (C-4), 132.0 (C-1), 121.8 (C-6), 117.6 (C-5), 116.8 (C-2), 104.8 (C-1′), 78.5 (C-5′), 78.3 ( C-3′), 75.6 (C-2′), 72.1 (C-4′), 72.0 (C-8), 63.2 (C-6′), 37.0 (C-7).22
Undatuside A (9) ‒ Colorless oil; 1H NMR (CD3OD, 400 MHz): δH 7.39 (2H, d, H-3, H-7), 7.32 (2H, t, H-4, H-6), 7.25 (1H, t, H-5), 4.85 (1H, d, J = 12.0 Hz, Ha-1), 4.63 (1H, d, J = 7.5 Hz, H-1′), 4.62 (1H, d, J = 12.0 Hz, Hb-1), 4.48 (1H, dd, J = 11.8, 2.0 Hz, Ha-6′), 4.23 (1H, dd, J = 11.8, 5.7 Hz, Hb-6′), 3.24-3.46 (4H, m, H-2′, H-5′, H-3′, H-4′), 2.77 (1H, d, J = 15.6 Hz, Ha-2′′), 2.72 (1H, d, J = 15.6 Hz, Hb-2′′), 2.67 (2H, s, H-4′′), 1.38 (3H, s, 3′′-CH3); 13C NMR (CD3OD, 100 MHz): δC 172.4 (C-5′′, C-1′′), 138.8 (C-2), 129.2 (C-4, C-6), 129.1 (C-7, C-3), 128.7 (C-5), 103.2 (C-1′), 77.7 (C-3′), 75.9 (C-5′), 74.9 (C-2′), 71.8 (C-1), 71.5 (C-4′), 70.7 (C-3′′), 64.6 (C-6′), 46.3 (C-2′′), 45.9 (C-4′′), 27.7 (3′′-CH3).23
Ginnalin A (10) ‒ Colorless crystalline; 1H NMR (aceton-d6, 400 MHz): δH 7.13 (2H, s, H-2, H-6), 7.11 (2H, s, H-2′′, H-6′′), 4.87 (1H, dt, J = 10.0, 5.5 Hz, H-2′), 4.56 (1H, d, J = 12.0 Hz, Ha-6′), 4.35 (1H, dd, J = 12.0, 5.0 Hz, Hb-6′), 4.05 (1H, dd, J = 10.5, 5.5 Hz, Ha-1′), 3.79 (1H, t, J = 9.0 Hz, H-3′), 3.59‒3.54 (2H, m, H-4′, H-5′), 3.34 (1H, t, J = 10.5 Hz, Hb-1′); l3C NMR (acetone-d6, 100 MHz): δC 168.8 (C-7), 166.5 (C-7′′), 145.9 (C-3, C-5), 145.8 (C-3′′, C-5′′), 138.9 (C-4), 138.7 (C-4′′), 121.5 (C-1), 121.2 (C-1′′), 109.7 (C-2, C-6), 109.3 (C-2′′, C-6′′), 79.4 (C-5′), 76.4 (C-3′), 72.8 (C-2′), 71.5 (C-4′),64.5 (C-6′), 67.4 (C-1′).24
5β, 8β-syrretaglucone C (11) ‒ White amorphous powder; 1H NMR (CD3OD, 400 MHz): δH 7.51 (1H, s, H-3), 6.67 (1H, d, J = 8.0 Hz, H-5′), 6.65 (1H, d, J = 1.9 Hz, H-2′), 6.58 (1H, s, H-1), 6.53 (1H, dd, J = 8.0, 1.9 Hz, H-6′). 4.17 (2H, dt, J = 14.2, 7.1 Hz, H-8′), 3.81 (1H, q, J = 6.5 Hz, H-9), 3.73 (1H, overlapped, H-5), 3.71 (3H, s, H-13), 3.17 (3H, s, H-11), 2.75 (2H, t, J = 7.1 Hz, H-7′), 2.61 (1H, dd, J = 15.1, 5.5 Hz, Ha-6), 2.47 (1H, dd, J = 15.1, 4.4 Hz, Hb-6), 1.28 (3H, d, J = 6.5 Hz, H-10); 13C NMR (CD3OD, 100 MHz): δC 174.0 (C-7), 169.3 (C-12), 153.8 (C-3), 147.1 (C-3′), 145.8 (C-4′), 141.3 (C-1), 131.7 (C-1′), 122.0 (C-6′), 119.4 (C-8), 117.8 (C-2′), 117.2 (C-5′), 111.1 (C-4), 78.9 (C-9), 67.4 (C-8′), 56.8 (12-OCH3), 52.9 (9-OCH3), 42.1 (C-6), 36.2 (C-7′), 28.7 (C-5), 19.2 (C-10).25
Loliolide (12) ‒ White amorphous powder; 1H NMR (DMSO-d6, 400 MHz): δH 5.81 (1H, s, H-7), 4.85 (1H, s, OH), 3.96 (1H, m, H-3), 2.34 (1H, dd, J = 11.5, 2.0 Hz, Ha-4), 1.86 (1H, dd, J = 12.6, 2.3 Hz, Ha-2), 1.53 (3H, s, H-11), 1.17 (1H, t, J = 11.5 Hz, Hb-4), 1.28 (1H, t, J = 12.6 Hz, Hb-2), 1.23 (3H, s, H-9), 1.18 (3H, s, H-10); 13C NMR (DMSO-d6, 100 MHz): δC 181.8 (C-6), 170.9 (C-8), 112.4 (C-7), 86.5 (C-5), 62.8 (C-3), 49.7 (C-2), 47.9 (C-4), 34.8 (C-1), 29.8 (C-10), 25.3 (C-11), 24.7 (C-9).26
3-(4′-hydroxyphenyl)-2-propenoic acid (4′′-carboxyl)-phenyl ester (13) ‒ Colorless needles; 1H NMR (acetone-d6, 400 MHz): δH 7.82 (2H, d, J = 8.6 Hz, H-3′, H-5′), 7.60 (1H, d, J = 15.5 Hz, H-3), 7.46 (2H, d, J = 8.6 Hz, H-2′, H-6′), 6.91 (2H, d, J = 8.6 Hz, H-2′′, H-6′′), 6.88 (2H, d, J = 8.6 Hz, H-3′′, H-5′′), 6.32 (1H, d, J = 15.5 Hz, H-2); 13C NMR (acetone-d6, 100 MHz): δC 168.0 (C-1), 167.3 (C-7′′), 162.4 (C-1′′), 160.4 (C-4′), 145.5 (C-3), 132.7 (C-2′, C-6′), 130.9 (C-2′′, C-6′′), 129.7 (C-4′′), 127.1 (C-1′), 116.6 (C-2), 115.8 (C-3′′, C-5′′), 115.7 (C-3′, C-5′).27
3-O-p-Coumaroylquinic acid (14) ‒ White amorphous powder; 1H NMR (CD3OD, 400 MHz): δH 7.68 (1H, d, J = 15.9 Hz, H-7), 7.48 (2H, d, J = 8.5 Hz, H-2, H-6), 6.82 (2H, d, J = 8.3 Hz, H-3, H-5), 6.38 (1H, d, J = 15.9 Hz, H-8), 5.38 (1H, m, H-3′), 4.11 (1H, m, H-5′), 3.72 (1H, dd, J = 7.6, 2.7 Hz, H-4′), 2.21–1.94 (4H, m, H-2′, H-6′); 13C NMR (CD3OD, 100 MHz): δC 177.6 (C-7′), 168.9 (C-9), 161.2 (C-4), 146.4 (C-7), 131.1 (C-2, C-6), 127.4 (C-1), 116.8 (C-3, C-5), 115.9 (C-8), 76.4 (C-1′), 74.2 (C-4′), 72.6 (C-3′), 68.9 (C-5′), 36.9 (C-2′), 36.2 (C-6′).28
Cryptochlorogenic acid methyl ester (15) ‒ White amorphous powder; 1H NMR (CD3OD, 400 MHz): δH 7.65 (1H, d, J =15.9 Hz, H-7), 7.06 (1H, d, J = 1.6 Hz, H-2), 6.99 (1H, dd, J = 8.2, 1.6 Hz, H-5), 6.80 (1H, d, J = 8.2 Hz, H-6), 6.38 (d, J = 15.9 Hz, H-8), 4.84 (dd, J = 8.6, 3.5 Hz, H-4′), 4.29 (d, J = 3.5 Hz, H-5′), 4.27 (d, J = 3.5 Hz, H-3′), 3.73 (3H, s, -OCH3), 2.20 (1H, m, Ha-6′), 2.04 (1H, m, Hb-6′), 2.24 (1H, m, Ha-2′), 2.05 (1H, m, Hb-2′); 13C NMR (CD3OD, 100 MHz): δC 175.6 (C-7′), 168.9 (C-9), 149.6 (C-4), 147.1 (C-7), 146.8 (C-3), 127.5 (C-1), 122.9 (C-6), 116.6 (C-5), 115.3 (C-8), 115.2 (C-2), 7б.4 (C-1′), 78.5 (C-4′), 69 (C-5′), 65.6 (C-3′), 52.9 (-OCH3), 42.1 (C-2′), 37.7 (C-6′).29
Osmanthuside B (16) ‒ White amorphous powder; 1H NMR (CD3OD, 400 MHz): δH 7.67 (1H. d. J = 16.1 Hz, H-7), 7.46 (2H, d, J = 8.6 Hz, H-2, H-6), 7.07 (2H, d, J = 8.6 Hz, H-2′, H-6′), 6.81 (2H. d, J = 8.6 Hz, H-3, H-5), 6.72 (2H, d, J = 8.6 Hz, H-3′, H-5′), 6.34 (1H. d, J = 16.1 Hz, H-8), 5.22 (1H, d, J = 1.6 Hz, H-1′′′), 4.91 (1H, t, J = 8.1 Hz, H-1′′), 4.38 (1H, d, J = 8.1 Hz, H-4′′), 4.11 (1H, m, H-2′′), 3.90 (1H, m, H-3′′), 3.85 (1H, dd, J = 12.0, 2.0 Hz, Ha-6′′), 3.74 (2H, m, H-8′), 3.73 (1H, m, H-5′′), 3.65 (1H, dd, J = 12.0, 6.0 Hz, Hb-6′′), 3.62 (1H, t. J = 8.1 Hz, H-4′′′), 3.57(1H. m, H-5′′′), 3.47(1H. t, J = 8.1 Hz, H-2′′′), 3.41 (1H. t, J = 9.2 Hz, H-3′′′), 2.84 (2H, t, J = 6.8 Hz, H-7′), 1.10 (3H. d, J = 6.1 Hz, H-6′′′); 13C NMR (CD3OD, 100 MHz): δC 168.2 (C-9), 161.3 (C-4), 156.8 (C-4′), 147.6 (C-7), 131.3 (C-2, C-6), 130.9 (C-2′, C-6′), 130.6 (C-1′), 127.0 (C-1), 116.8 (C-3, C-5), 116.1 (C-3′, C-5′), 114.7 (C-8), 104.1 (C-1′′), 102.9 (C-1′′′), 81.6 (C-3′′), 76.1 (C-5′′′), 75.9 (C-5′′), 73.8 (C-2′′′), 72.3 (C-8′), 72.2 (C-2′′), 72.0 (C-3′′′), 70.6 (C-4′′′), 70.3 (C-4′′), 62.3 (C-6′′), 36.3 (C-7′), 18.4 (C-6′′′).30
Magnoloside A (17) ‒ White amorphous powder; 1H NMR (DMSO-d6, 400 MHz): δH 7.62 (1H, d, J = 16.0 Hz, H-7), 7.09 (1H, d, J = 2.0 Hz, H-2), 6.97 (1H, dd, J = 8.5, 2.0 Hz, H-6), 6.81 (1H, d, J = 8.5 Hz, H-5′), 6.72 (1H, d, J = 2.0 Hz, H-2′), 6.68 (1H, d, J = 8.0 Hz, H-5), 6.58 (1H, dd, J = 8.0, 2.0 Hz, H-6′), 6.36 (1H, d, J = 16.0 Hz, H-8), 5.18 (1H, d, J = 2.3 Hz, H-1′′′), 4.95 (1H, overlapped, H-4′′), 4.35 (1H, d, J = 8.0 Hz, H-1′′), 4.01 (1H, m, Ha-8′), 3.94 (1H, m, H-3′′′), 3.81 (1H, m, H-3′′), 3.68 (1H, m, Hb-8′), 3.50‒3.71 (5H, m, H-2′′′, H-5′′, H-5′′′, Ha-6′′, Hb-6′′), 3.39 (1H, m, H-2′′), 3.31 (1H, m, H-4′′′), 2.79 (2H, t, J = 7.2 Hz, H-7′), 1.09 (3H, d, J = 6.4 Hz, H-6′′′); 13C NMR (DMSO-d6, 100 MHz): δC 168.9 (C-9), 149.4 (C-4′), 147.2 (C-7), 146.3 (C-3′), 145.8 (C-3), 144.4 (C-4), 131.6 (C-1), 127.5 (C-1′), 123.2 (C-6′), 121.3 (C-6), 117.1 (C-2), 116.5 (C-5′), 116.4 (C-5), 115.1 (C-2′), 114.8 (C-8), 100.6 (C-1′′), 98.3 (C-1′′′), 75.8 (C-5′′), 73.8 (C-4′′′), 73.5 (C-2′′), 72.1 (C-3′′′), 71.9 (C-2′′′), 71.8 (C-7′), 71.1 (C-3′′), 69.8 (C-5′′′), 66.8 (C-4′′), 62.6 (C-6′′), 36.5 (C-8′), 17.7 (C-6′′′).31
Echinacoside (18) ‒ White amorphous powder; 1H NMR (CD3OD, 400 MHz): δH 7.58 (1H, d, J = 19.1 Hz, H-7), 7.06 (1H, d, J = 2.1 Hz, H-2), 6.93 (1H, dd, J = 8.3, 2.1 Hz, H-6), 6.76 (1H, d, J = 8.3 Hz, H-5′), 6.69 (1H, d, J = 2.1 Hz, H-2′), 6.68 (1H, d, J = 8.5 Hz, H-5), 6.57 (1H, dd, J = 8.5, 2.1 Hz, H-6′), 6.26 (1H, d, J = 19.1 Hz, H-8), 5.16 (1H, d, J = 1.2 Hz, H-1′′′), 5.01 (1H, t, J = 9.5 Hz, H-4′′, H-1′′′′), 4.37 (1H, d, J = 9.5 Hz, H-1′′), 3.99 (1H, m, Ha-8′), 3.71‒3.97 (10H, m-overlapped, H-3′′, H-3′′′, H-3′′′′, Ha-6′′, Hb-6′′, H-5′′, H-5′′′, H-5′′′′, Ha-6′′′′, Hb-6′′′′), 3.70 (1H, m, Hb-8′), 3.47‒3.54 (5H, m-overlapped, H-2′′, H-2′′′, H-2′′′′, H-4′′′, H-4′′′′), 2.78 (2H, t, J = 5.7 Hz, H-7′), 1.06 (3H, d, J = 7.4 Hz, H-6′′′); 13C NMR (CD3OD, 100 MHz): δC 168.5 (C-9), 149.8 (C-3), 148.4 (C-7), 146.8 (C-4), 146.2 (C-4′), 144.8 (C-3′), 131.6 (C-1), 127.4 (C-1′), 123.2 (C-6), 121.2 (C-6′), 117.3 (C-5′), 116.6 (C-2′), 116.4 (C-5), 115.4 (C-8), 114.7 (C-2), 104.8 (C-1′′′), 104.3 (C-1′′′′), 103.2 (C-1′′), 81.7 (C-3′′), 78.1 (C-3′′′′), 77.9 (C-5′′′′), 76.1 (C-2′′), 74.9 (C-5′′, C-2′′′′), 73.9 (C-4′′′), 72.5 (C-8′, C-2′′′), 72.2 (C-3′′′), 71.6 (C-4′′′′), 70.5 (C-4′′, C-5′′′), 69.5 (C-6′′), 62.7 (C-6′′′′), 36.5 (C-7′), 18.5 (C-6′′′).32
Jasnervoside B (19) ‒ White amorphous powder; 1H NMR (CD3OD, 400 MHz): δH 7.67 (1H, d, J = 15.9 Hz, H-7), 7.61 (1H, d, J = 15.8 Hz, H-7′), 7.47 (2H, d, J = 8.6 Hz, H-2′, H-6′), 7.07 (1H, d, J = 1.9 Hz, H-2), 6.96 (1H, dd, J = 8.2, 1.9 Hz, H-6), 6.81 (2H, d, J = 8.6 Hz, H-3′, H-5′), 6.77 (1H, d, J = 8.2 Hz, H-6′′), 6.71 (1H, d, J = 8.2 Hz, H-5), 6.68 (1H, d, J = 8.0 Hz, H-5′′), 6.59 (1H, dd, J = 1.9 Hz, H-2′′), 6.39 (1H, d, J = 15.9 Hz, H-8), 6.30 (1H, d, J = 15.8 Hz, H-8′), 5.20 (1H, d, J = 1.3 Hz, H-1′′′′′), 5.10 (1H, dd, J = 9.8, 3.3 Hz, H-3′′′), 4.99 (2H, m, H-4′′′, H-4′′′′′), 4.90 (1H, brs, H-1′′′′), 4.40 (1H, d, J = 7.9 Hz, H-1′′′), 4.11 (1H, m, Ha-8′′), 3.85‒3.78 (2H, m-overlapped, Hb-8′′, H-2′′′), 3.78‒3.67 (2H, m-overlapped, Ha-6′′′, H-5′′′), 3.60 (2H, m-overlapped, H-5′′′′, H4′′′′), 3.59 (1H, m, H-2′′′′), 3.58 (2H, m-overlapped, H-3′′′′, H-2′′′′′), 3.55 (1H, m, H-3′′′′′), 3.54 (1H, m, H-5′′′′′), 3.52 (1H, m, Hb-6′′′), 2.81 (2H, td, J = 7.3, 4.3 Hz, H-7′′), 1.25 (3H, d, J = 6.2 Hz, H-6′′′′), 1.21 (3H, d, J = 6.2 Hz, H-6′′′′′); 13C NMR (CD3OD, 100 MHz): δC 169.9 (C-9′), 168.1 (C-9), 161.2 (C-4′), 149.7 (C-7′), 148.1 (C-7), 146.9 (C-3), 146.7 (C-4), 146.1 (C-4′′), 144.7 (C-3′′), 131.4 (C-1′′), 131.2 (C-2′, C-6′), 127.6 (C-1), 127.2 (C-1′), 123.3 (C-6), 121.3 (C-6′′), 117.1 (C-2), 116.8 (C-3′, C-5′), 116.4 (C-2′′), 116.3 (C-5′′), 116.1 (C-5), 115.3 (C-8′), 114.7 (C-8), 104.3 (C-1′′′′), 103.1 (C-1′′′′′), 102.3 (C-1′′′), 81.6 (C-3′′′), 76.1 (C-2′′′), 75.4 (C-3′′′′), 74.6 (C-5′′′), 73.8 (C-4′′′), 73.7 (C-4′′′′′), 72.5 (C-8′′), 72.4 (C-5′′′′), 72.3 (C-2′′′′), 72.1 (C-2′′′′′), 72.0 (C-3′′′′′), 71.4 (C-4′′′′), 70.4 (C-5′′′′′), 67.9 (C-6′′′), 36.6 (C-7′′), 18.5 (C-6′′′′), 18.1 (C-6′′′′′).33
Pedunculoside (20) ‒ White amorphous powder; 1H NMR (CD3OD, 400 MHz): δH 5.36 (1H, br.s, H-1′), 5.34 (1H, br.s, H-12), 3.82 (1H, dd, J = 12.0, 2.0 Hz, Ha-6′), 3.71 (1H, dd, J = 12.0, 4.5 Hz, Hb-6′), 3.64 (1H, dd, J = 11.5, 4.5 Hz, H-3), 3.56 (1H, d, J = 10.5 Hz, Ha-23), 3.43 (1H, m, H-5′), 3.39 (1H, m, H-4′), 3.36 (1H, m, H-2′), 3.35 (1H, m, H-3′), 3.32 (1H, d, J = 10.5 Hz, Hb-23), 2.63 (1H, dt, J = 11.5, 4.5 Hz, Ha-16), 2.54 (1H, br.s, H-18), 2.1 (2H, m, H-11), 1.87 (1H, m, Ha-15), 1.81 (1H, m, Ha-22), 1.76 (1H, m, Ha-21), 1.75 (1H, m, H-9), 1.68 (2H, m, H-1, Ha-7), 1.66 (2H, m, Hb-16, Hb-22), 1.65 (2H, m, H-2), 1.47 (2H, m, H-6), 1.38 (1H, m, H-20), 1.36 (3H, s, H-27), 1.32 (1H, m, Hb-7), 1.29 (1H, m, Hb-21), 1.21 (3H, s, H-29), 1.19 (1H, m, H-5), 1.03 (2H, m, H-1), 1.01 (3H, s, H-25), 1.01 (1H, m, Hb-15), 0.96 (3H, d, J = 7.0 Hz, H-30), 0.81 (3H, s, H-26), 0.72 (3H, s, H-24); 13C NMR (CD3OD, 100 MHz): δC 177.8 (C-28); 140.1 (C-13), 129.3 (C-12), 95.7 (C-1′), 80.1 (C-5′), 79.8 (C-3′), 75.1 (C-2′), 74.1 (C-3), 73.6 (C-19), 72.1 (C-4′), 67.5 (C-23), 62.4 (C-6′), 55.1 (C-18), 49.4 (C-17), 49.1 (C-5), 48.4 (C-9), 43.2 (C-4), 42.8 (C-20), 42.6 (C-14), 41.1 (C-8), 39.5 (C-1), 38.2 (C-22), 37.8 (C-10), 33.6 (C-7), 29.6 (C-15), 27.5 (C-2), 27.1 (C-21), 27.2 (C-29), 26.6 (C-16), 24.6 (C-27), 24.6 (C-11), 19.2 (C-6), 17.8 (C-26), 16.7 (C-30), 16.5 (C-25), 12.6 (C-24).34
PTP1B inhibitory assay ‒ In the PTP1B inhibitory assay, the reaction buffer containing 50 mM citrate (pH 6.0), 1 mM ethylene diamine tetraacetic acid, 0.1 M NaCl, and 1 mM dithiothreitol was prepared.35 Then, 10 μL PTP1B enzyme diluted using 30 μL of the reaction buffer was added to each well of a 96-well plate (final volume 100 μL), with or without a test compound/control with a concentration range of 1-100 μM. Then, 50 μL of p-NPP substrate (2 mM) dissolved in PTP1B reaction buffer was added. The reaction mixture was incubated for 15min in the dark at 37oC, and then the reaction was terminated by the addition of 10 μL of NaOH (10 M). The amount of p-nitrophenolate, which was produced by enzymatic dephosphorylation of the p-NPP substrate was measured at 405nm wavelength using a VERSA max microplate spectrophotometer (Molecular Devices, Sunnyvale, CA, USA). In the absence of the PTP1B enzyme, the non-enzymatic hydrolysis of the p-NPP substrate (2 mM) was corrected by measuring the increase in absorbance at 405 nm wavelength.35 Then, the percent inhibition (%) was calculated as follows: % inhibition = {(Ac−As) / Ac} × 100, where Ac and As are the absorbance of the control and sample, respectively.35 In this assay, based on the previous disclosure of ursolic acid and its derivatives' strong inhibitory potential against PTP1B,36 this compound has been selected as a positive control. The concentration capable of inhibiting 50% of the enzymatic activity (IC50, μM) was represented for the inhibitory activity of tested compounds.35
α-Glucosidase inhibitory assay ‒ According to the previously published procedure with a slight modification, the α-glucosidase enzyme was prepared at 0.1 units by dissolving with 10 mM phosphate buffer, then treated with the isolated compounds at various concentrations (1‒50 µM). The reaction was performed in 96-well plates using 2.5mM p-nitrophenyl α-D-glucopyranoside (p-NPG) as the substrate in 100 mM phosphate buffer (pH = 6.8) at 37oC. After 10 min, the hydrolyzation of p-NPG released p-nitrophenol, which generated a yellow anion under alkaline conditions, allowing simple spectrophotometric quantitation of α-glucosidase activity, which can be measured at 405 nm.35 Considering the prior announcement regarding the potent inhibition of α-glucosidase by acarbose, this substance has been designated as a positive control.37
Enzyme kinetic analysis with PTP1B and α-glucosidase ‒ To study the inhibition mode of PTP1B, various concentrations of p-NPP substrate were utilized, both in the presence and absence of different concentrations of the test compound. Similarly, for α-glucosidase inhibition was obtained in the presence of various concentrations of the p-NPG substrate. Lineweaver-Burk double reciprocal plots were constructed to analyze the enzyme’s kinetic parameters, such as Km and Vmax. The inhibition constant (Ki) was determined by interpreting Dixon plots, where the x-axis value represented Ki. This approach provided valuable insights into the inhibition mode of PTP1B and α-glucosidase.35
Statistics ‒ Statistical comparison between groups was analyzed using one-way ANOVA and Duncan’s test (Systat Inc., Evanston, IL, USA). Data are expressed as the mean ± SEM (standard error of the mean) of four independent experiments. Values of p < 0.05 were considered statistically significant.
Results and Discussion
Compound 10 was isolated as a colorless, crystalline product. The 1H and 13C NMR spectra of 10 showed typical signals of two units of galloyl moieties (two trihydroxybenzoyl groups) at δH/δC 7.13 (2H, s, H-2, H-6)/109.7 and 7.11 (2H, s, H-2′′, H-6′′)/109.3; six hydroxylated aromatic carbons at δC 145.9 (C-3, C-5), 145.8 (C-3′′, C-5′′), 138.9 (C-4), and 138.7 (C-4′′); two quaternary carbon signals at δC 121.5 (C-1) and 121.2 (C-1′′); combine with two carboxyl groups at δC 168.8 (C-7) and 166.5 (C-7′′). In addition, the remaining signals belong to a 1,5-anhydroglucitol moiety at δH 4.87 (1H, dt, J = 10.0, 5.5 Hz, H-2′), 4.56 (1H, d, J = 12.0 Hz, Ha-6′), 4.35 (1H, đ, J = 12.0, 5.0 Hz, Hb-6′), 4.05 (1H, dd, J = 10.5, 5.5 Hz, Ha-1′), 3.79 (1H, t, J = 9.0 Hz, H-3′), 3.59‒3.54 (2H, m, H-4′, H-5′), and 3.34 (1H, t, J = 10.5 Hz, Hb-1′) and δC 79.4 (C-5′), 76.4 (C-3′), 72.8 (C-2′), 71.5 (C-4′), 67.4 (C-1′), and 64.5 (C-6′). The downfield shift signals of H-2′ and two H-6′ suggested that the two galloyl groups in 10 were attached to the positions 2′ and 6′ via two ester linkages. Based on the above analysis and comparison with the literature, compound 10 was identi- fied as ginnalin A (Fig. 1).24
Compound 13 was obtained as colorless needles. The 1H and 13C NMR spectra of 13 showed signals for eight aromatic protons of two AABB coupling systems at δH/δC 7.82 (2H, d, J = 8.6 Hz, H-3′, H-5′)/132.7, 7.46 (2H, d, J = 8.6 Hz, H-2′, H-6′)/115.7, 6.91 (2H, d, J = 8.6 Hz, H-2′′, H-6′′)/130.9, 6.88 (2H, d, J = 8.6 Hz, H-3′′, H-5′′)/115.8; one trans-double bond at δH/δC 7.60 (1H, d, J = 15.5 Hz, H-3)/145.5 and 6.32 (1H, d, J = 15.5 Hz, H-2)/116.6; together with two carbons at δC 168.0 (C-1) and 167.3 (C-7′′). In addition, the HMBC showed correlations from H-2 (δH 6.32)/H-3 (δH 7.60) to C-1′ (δC 127.1) and C-1 (δC 168.0); as well as from H-3′′, H-5′′ (δH 6.88) to C-7′′ (δC 167.3) suggested the presence of 3-(4′-hydroxyphenyl)-2-propenoic acid and 4′′-carboxyl-phenol moieties, and these moieties connected to each other via a ester linkage. Based on the above analysis and comprehensive comparison with relevant literature, compound 13 was determined to be 3-(4′-hydroxyphenyl)-2-propenoic acid (4′′-carboxyl)-phenyl ester (Fig. 1).27
Based on NMR spectroscopic analysis and comparison with the reported data in the pieces of literature, the remain isolated compounds were identified as staphylionoside H (1),15 phaseic acid (2),16 dehydrovomifoliol (3),17 dehydrovomifoliol glycoside (4),18 icariside E5 (5),19 saikolignanoside A (6),20 salidroside (7),21 3,4-dihydroxyphenylethyl alcohol 8-O-β-D-glucopyranoside (8),22 undatuside A (9),23 5β, 8β-syrretaglucone C (11),25 loliolide (12),26 3-O-p-coumaroylquinic acid (14),28 cryptochlorogenic acid methyl ester (15),29 osmanthuside B (16),30 magnoloside A (17),31 echinacoside (18),32 jasnervoside B (19),33 and pedunculoside (20)34 (Fig. 1).
In the treatment of metabolic disorders like diabetes and obesity, targeting PTP1B and α-glucosidase enzymes has shown significant promise. In vitro assays were performed to assess the inhibitory potential of isolated compounds against these enzymes. The results, presented in Table 1, demonstrate that compounds 1‒20 exhibited varying levels of PTP1B inhibition in a dose-dependent manner, with IC50 values ranging from 7.9 to 94.5 μM. Notably, compound 13 (IC50 = 7.9 μM) exhibited strong PTP1B inhibition and compound 10 with IC50 of 37.6 μM, while the argument was the positive control [ursolic acid (IC50 = 5.8 μM)]. Moreover, compounds 10 and 13 demonstrated remarkable activity against α-glucosidase, with IC50 values of 13.9 and 31.9 μM, respectively. These values were substantially higher than the positive control acarbose (IC50 = 289.5 μM). Interestingly, these two compounds, known for being the most potent PTP1B inhibitors, also displayed outstanding inhibition of α-glucosidase (Table 1).

The inhibitory activity of isolated compounds 1‒20 from H. cordata was evaluated against PTP1B and α-glucosidase enzymes
The enzyme inhibition properties of compounds 10 and 13 were analyzed using Lineweaver-Burk and Dixon plots. The graphical representation in Figs. 2 and 3 clearly showed that both compounds displayed non-competitive PTP1B and α-glucosidase inhibition, as indicated by the intersection of their regression lines on the left side of the y-axis. The inhibition constants (Ki) of 10 and 13 were determined from the Dixon plots with values of 35.6 μM and 7.3 μM for PTP1B and 13.9 μM and 31.0 μM for α-glucosidase, respectively (Table 1). These findings provide valuable insights into the inhibitory behaviour of these compounds and their potential in enzyme-targeted therapeutic applications.
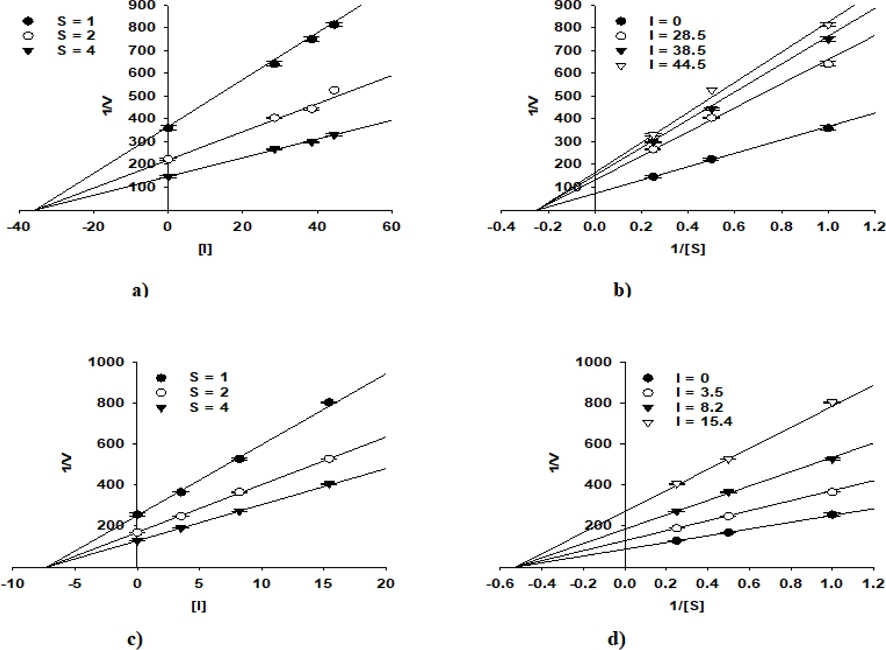
Dixon plots for the PTP1B inhibition of compounds 10 (a) and 13 (c). Lineweaver-Burk plots for the PTP1B inhibition of compounds 10 (b) and 13 (d).
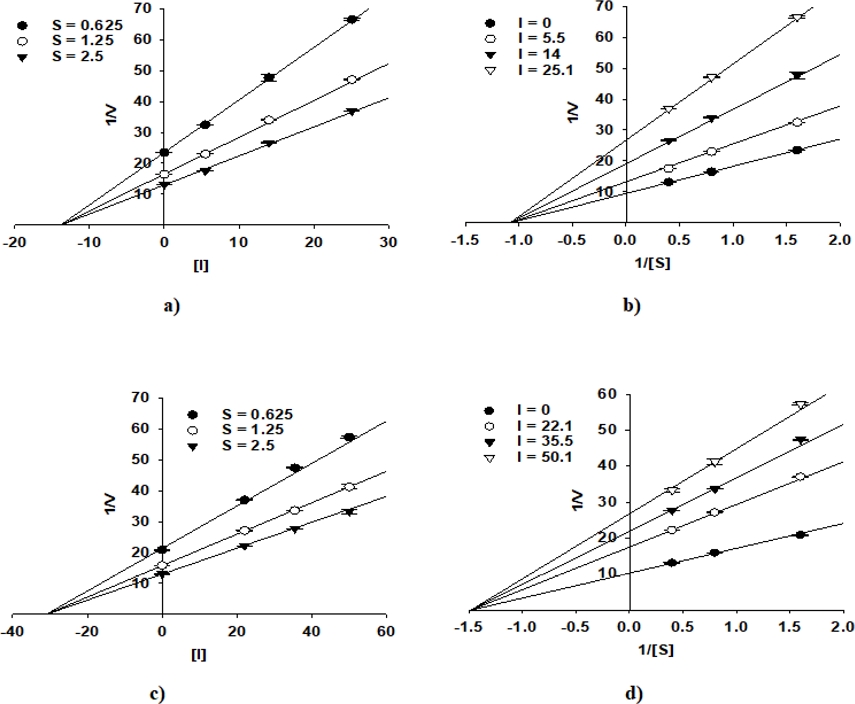
Dixon plots for α-glucosidase inhibition of 10 (a) and 13 (c). Lineweaver-Burk plots for α-glucosidase inhibition of 10 (b) and 13 (d).
In conclusion, a total of 20 compounds were isolated from the whole plants of H. cordata, encompassing various megastigmanes, phenolics, coumarins, and other derivatives. The identification of these compounds was confirmed through NMR spectroscopic analysis and comparison with existing literature data. Among them, compounds 13 and 10 demonstrated strong inhibition against PTP1B, with IC50 values of 7.9 μM and 37.6 μM, respectively. They also displayed significant inhibition against α-glucosidase, with IC50 values of 13.9 μM for compound 10 and 31.9 μM for compound 13, respectively. The calculated PTP1B inhibition constants (Ki) were 35.6 μM for compound 10 and 7.3 μM for compound 13, while the α-glucosidase inhibition constants (Ki) were 13.9 μM for compound 10 and 31.0 μM for compound 13, respectively. These findings emphasize the potential therapeutic value of compounds 10 and 13 in addressing metabolic disorders, making them promising candidates for further investigation and potential development as treatments for conditions like diabetes and obesity.
Acknowledgments
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT (2021R1A2C2011940), Korea. The authors would like to thank the Korea Basic Science Institute (KBSI) for mass spectrometric measurements.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
-
Sun, H.; Saeedi, P.; Karuranga, S.; Pinkepank, M.; Ogurtsova, K.; Duncan, B. B.; Stein, C.; Basit, A.; Chan, J. C. N.; Mbanya, J. C.; Pavkov, M. E.; Ramachandaran, A.; Wild, S. H.; James, S.; Herman, W. H.; Zhang, P.; Bommer, C.; Kuo, S.; Boyko, E. J.; Magliano, D. J. Diabetes Res. Clin. Pract. 2022, 183, 109119.
[https://doi.org/10.1016/j.diabres.2021.109119]

-
Banday, M. Z.; Sameer, A. S.; Nissar, S. Avicenna J. Med. 2020, 10, 174–188.
[https://doi.org/10.4103/ajm.ajm_53_20]

-
Galicia-Garcia, U.; Benito-Vicente, A.; Jebari, S.; Larrea-Sebal, A.; Siddiqi, H.; Uribe, K. B.; Ostolaza, H.; Martín, C. Int. J. Mol. Sci. 2020, 21, 6275.
[https://doi.org/10.3390/ijms21176275]

-
Lebovitz, H. E. Endocrinol. Metab. Clin. North Am. 1997, 26, 539–551.
[https://doi.org/10.1016/S0889-8529(05)70266-8]

-
Rebollar-Ramos, D.; Ovalle-Magallanes, B.; Palacios-Espinosa, J. F.; Macías-Rubalcava, M. L.; Raja, H. A.; González-Andrade, M.; Mata, R. ACS Omega 2021, 6, 22969–22981.
[https://doi.org/10.1021/acsomega.1c03708]

- Jiangang, F.; Ling, D.; Zhang, L.; Hongmei, L. Chin. Med. 2013, 4, 101–123.
-
Shim, S.-Y.; Seo, Y.-K.; Park, J.-R. J. Med. Food 2009, 12, 383–388.
[https://doi.org/10.1089/jmf.2007.0684]

-
Chiang, L.-C.; Chang, J.-S.; Chen, C.-C.; Ng, L.-T.; Lin, C.-C. Am. J. Chin. Med. 2003, 31, 355–362.
[https://doi.org/10.1142/S0192415X03001090]

-
Bauer, R.; Pröbstle, A.; Lotter, H.; Wagner-Redecker, W.; Matthiesen, U. Phytomedicine 1996, 2, 305–308.
[https://doi.org/10.1016/S0944-7113(96)80073-0]

-
Hayashi, K.; Kamiya, M.; Hayashi, T. Planta Med. 1995, 61, 237–241.
[https://doi.org/10.1055/s-2006-958063]

-
Lu, H. M.; Liang, Y. Z.; Yi, L. Z.; Wu, X. J. J. Ethnopharmacol. 2006, 104, 245–249.
[https://doi.org/10.1016/j.jep.2005.09.012]

-
Chen, Y.-Y.; Liu, J.-F.; Chen, C.-M.; Chao, P.-Y.; Chang, T.-J. J. Nutr. Sci. Vitaminol. 2003, 49, 327–333.
[https://doi.org/10.3177/jnsv.49.327]

-
Choi, C. W.; Kim, S. C.; Hwang, S. S.; Choi, B. K.; Ahn, H. J.; Lee, M. Y.; Park, S. H.; Kim, S. K. Plant Sci. 2002, 163, 1161–1168.
[https://doi.org/10.1016/S0168-9452(02)00332-1]

-
Kim, S. K.; Ryu, S. Y.; No, J.; Choi, S. U.; Kim, Y. S. Arch. Pharm. Res. 2001, 24, 518–521.
[https://doi.org/10.1007/BF02975156]

-
Yu, Q.; Matsunami, K.; Otsuka, H.; Takeda, Y. Chem. Pharm. Bull. 2005, 53, 800–807.
[https://doi.org/10.1248/cpb.53.800]

-
Hirai, N.; Kondo, S.; Ohigashi, H. Biosci. Biotech. Biochem. 2003, 67, 2408–2415.
[https://doi.org/10.1271/bbb.67.2408]

-
Takasugi, M.; Anetai, M.; Katsui, N.; Masamune, T. Chem. Lett. 1973, 2, 245–248.
[https://doi.org/10.1246/cl.1973.245]

-
Khan, S. H.; Mosihuzzaman, M.; Nahar, N.; Rashid, M. A.; Rokeya, B.; Ali, L.; Azad Khan, A. K. Pharm. Biol. 2003, 41, 512–515.
[https://doi.org/10.1080/13880200308951345]

-
Iorizzi, M.; Lanzotti, V.; De Marino, S.; Zollo, F.; Blanco-Molina, M.; Macho, A.; Munoz, E. J. Agric. Food Chem. 2001, 49, 2022–2029.
[https://doi.org/10.1021/jf0013454]

-
Zheng, Z. P.; Li, Y.; Ma, Z. Q.; Wang, Y.; Wang, Q. Chem. Nat. Compd. 2022, 58, 782–783.
[https://doi.org/10.1007/s10600-022-03795-7]

-
Mao, Y.; Li, Y.; Yao, N. J. Pharm. Biomed. Anal. 2007, 45, 510–515.
[https://doi.org/10.1016/j.jpba.2007.05.031]

-
Park, H. J.; Lee, M. S.; Lee, K. T.; Sohn, I. C.; Han, Y. N.; Miyamoto, K. I. Chem. Pharm. Bull. 1999, 47, 1029–1031.
[https://doi.org/10.1248/cpb.47.1029]

-
Wu, X.; Wang, Y.; Huang, X. J.; Fan, C. L.; Wang, G. C.; Zhang, X. Q.; Zhang, Q. W.; Ye, W. C. J. Asian Nat. Prod. Res. 2011, 13, 728–733.
[https://doi.org/10.1080/10286020.2011.586944]

-
Hatano, T.; Hattori, S.; Ikeda, Y.; Shingu, T.; Okuda, T. Chem. Pharm. Bull. 1990, 38, 1902–1905.
[https://doi.org/10.1248/cpb.38.1902]

-
Guo, S.; Liu, Y.; Sun, Y. P.; Pan, J.; Guan, W.; Li, X. M.; Wang S. Y.; Algradi, A. M.; Yang, B. Y.; Kuang, H. X. Nat. Prod. Res. 2022, 36, 4957–4966.
[https://doi.org/10.1080/14786419.2021.1914031]

-
Yang, X.; Kang, M. C.; Lee, K. W.; Kang, S. M.; Lee, W. W.; Jeon, Y. J. Algae 2011, 26, 201–208.
[https://doi.org/10.4490/algae.2011.26.2.201]

-
Niu, F.; Cui, Z.; Chang, H. T.; Jiang, Y.; Chen, F. K.; Tu, P. F. Chin. J. Chem. 2006, 24, 1788–1791.
[https://doi.org/10.1002/cjoc.200690334]

-
Ma, F. Y.; Zhang, X. M.; Li, Y.; Zhang, M.; Tu, X. H.; Du, L. Q. Front. Nutr. 2022, 9, 970019.
[https://doi.org/10.3389/fnut.2022.970019]

-
Zhu, X.; Dong, X.; Wang, Y.; Ju, P.; Luo, S. Helv. Chim. Acta 2005, 88, 339–342.
[https://doi.org/10.1002/hlca.200590017]

-
He, Z. D.; Ueda, S.; Akaji, M.; Fujita, T.; Inoue, K.; Yang, C. R. Phytochemistry 1994, 36, 709–716.
[https://doi.org/10.1016/S0031-9422(00)89802-7]

-
Seo, K. H.; Lee, D. Y.; In, S. J.; Lee, D. G.; Kang, H. C.; Song, M. C.; Baek, N. I. Chem. Nat. Compd. 2015, 51, 660–665.
[https://doi.org/10.1007/s10600-015-1379-4]

-
Xie, J.; Deng, J.; Tan, F.; Su, J. J. Chromatogr. B Analyt. Technnol. Biomed. Life Sci. 2010, 878, 2665–2668.
[https://doi.org/10.1016/j.jchromb.2010.07.023]

-
Guo, Z. Y.; Li, P.; Huang, W.; Wang, J. J.; Liu, Y. J.; Liu, B.; Wang Y. L.; Wu S. B.; Kennelly, E. J.; Long, C. L. Phytochemistry 2014, 106, 124–133.
[https://doi.org/10.1016/j.phytochem.2014.07.011]

-
Hung, N. H. Vietnam J. Sci. Technol., 2016, 54 (4A), 221–226.
[https://doi.org/10.15625/2525-2518/54/4A/11997]

-
Kim, D. H.; Paudel, P.; Yu, T.; Ngo, T. M.; Kim, J. A.; Jung, H. A.; Yokozawa, T.; Choi, J. S. Chem. Biol. Interact. 2017, 278, 65–73.
[https://doi.org/10.1016/j.cbi.2017.10.013]

-
Zhang, W.; Hong, D.; Zhou, Y.; Zhang, Y.; Shen, Q.; Li, J. Y.; Hu, L. H.; Li, J. Biochim. Biophys. Acta-Gen. Subj. 2006, 1760, 1505–1512.
[https://doi.org/10.1016/j.bbagen.2006.05.009]

-
Martin, A. E.; Montgomery, P. A. Am. J. Health Syst. Pharm. 1996, 53, 2277–2290.
[https://doi.org/10.1093/ajhp/53.19.2277]