Effects of Coumarins from Roots of Paramignya scandens (Griff.) Craib on LPS-induced IL-1β and IL-10 Cytokine Production in RAW 264.7 Macrophages
Abstract
Based on our previous study, we evaluated the modulatory effects on LPS-induced IL-1β and IL-10 cytokine production in RAW 264.7 macrophages of several medicinal herbs, including P. scandens. The results showed that P. scandens extract showed significant effects on LPS-induced IL-1β and IL-10 cytokine production in RAW 264.7 macrophages. Therefore, in the current research, we focused on the P. scandens sample. Cytokine production effects bioassay-guided isolation of ethyl acetate fraction of 70% ethanol extract from roots of Paramignya scandens (XL) obtained seven coumarins (1–7). Their chemical structures were identified using spectroscopic methods (NMR and MS) and compared with those previously published data to be xanthyletin (1), luvangetin (2), clausenidin (3), nordentatin (4), dentatin (5), clausarin (6), and anisocoumarin E (7). This study represents the first report on the presence of compounds 3, 6, and 7 in the Paramignya genus and compounds 1 and 2 in XL. All isolates (1–7) exhibited significant inhibition of LPS-induced interleukin (IL)-1β production compared to the LPS 5 ng/mL control group, with IL-1β concentrations ranging from 42.77 to 69.76 pg/mL. Additionally, the IL-10 production induced by compounds 1–7 in LPS-stimulated RAW 264.7 macrophages ranged from 175.98 to 321.56 pg/mL, demonstrating a marked increase as compared to the LPS 5 ng/mL control group. The stimulatory effect on IL-10 production and inhibitory effect on IL-1β production of compounds 1, 2, and 6 gradually increased with the test concentration in both RAW 264.7 macrophages and LPS-induced RAW 264.7 macrophages. Compounds 1, 2, and 6 inhibited IL-1β production in LPS-induced RAW 264.7 macrophages with IC50 values of 10.70 ± 1.18 μM, 8.57 ± 1.05 μM, and 17.43 ± 1.05 μM, respectively. These findings highlight the potential of all the compounds derived from P. scandens roots in inducing IL-1β and IL-10 cytokines activity in LPS-stimulated RAW 264.7 macrophages. The results contributed to expanding the knowledge of the chemistry and bioactivities of P. scandens and provided valuable data for future investigations on this species.
Keywords:
Paramignya scandens, Coumarin, Interleukine-1β, Interleukine-10, Modulation, RAW 264.7 macrophagesIntroduction
In recent years, the world has had to deal with the emergence of dangerous infectious diseases. The immune system is the body’s natural defense, so improving the immune system is an urgent issue and deserves attention, especially in diseases related to immunodeficiency. The immunological responses include many mechanisms, including those involving the cytokines interleukin (IL)- 1β and IL-10.1 IL-1β is a pro-inflammatory cytokine that plays a crucial role in host defense responses to infection and injury.2 It not only promotes the production of extracellular matrix enzymes but also acts as a bridge between innate and adaptive immunity.3,4 On the other hand, IL-10 is an essential cytokine with potent anti-inflammatory properties and regulates the functions of various immune macrophages.5
IL-10 is encoded by the IL-10 gene, which is located on chromosome 1 and comprises 5 exons in humans.6 Numerous studies showed the anti-inflammatory effects of IL-10 via inhibiting of proinflammatory cytokines and chemokines: tumor necrosis factor (TNF), interferongamma (IFN-γ), IL-1α/β, and others.7 Moreover, IL-10 was demonstrated to increase the secretion of anti-inflammatory molecules such as IL-1 receptor antagonist, soluble TNF receptor, as well as IL-27, which participate and contribute to an anti-inflammatory milieu.8 Interestingly, both endogenous and exogenous IL-10 inhibited IL-1β in the microglia exposed to epileptogenic injury caused by STAT-3-dependent inhibition of NOD-like receptor family pyrin domain containing 3 (NLRP3) inflammasome activity. 9 Furthermore, Gao et al. presented that the levels of NLRP3, pro-caspase-1, pro-IL-1β, active caspase-1, and mature IL-1β in the midbrain were much higher in IL-10−/− mice than wildtype mice after 24 h of lipopolysaccharide (LPS) injection.10 These results were explained by IL-10−/− microglia producing more intracellular reactive oxygen species (iROS) and showed more profound activation of NADPH oxidase (NOX2) than wildtype microglia. In addition, the inhibition of NOX2-derived iROS production suppressed LPS-elicited caspase-1 activation and IL-1β maturation in IL-10−/− microglia in vitro and in vivo.8,10,11
Natural products play a crucial role in the development of new drugs for cytokine production effects and serve as lead compounds. Paramignya scandens (Griff.) Craib (XL) is a small woody liana or creeper belonging to the Rutaceae family.12 This species is mainly distributed in Ha Nam, Quang Tri, and Lam Dong, Vietnam.13 In traditional medicine, P. scandens is known for its diuretic effect and is often used to treat conditions such as hematuria, abdominal pain, flatulence, and indigestion.13 Despite its traditional use, there has been limited research on the chemical compositions and biological effects of P. scandens. However, some studies have reported the presence of tirucallanes, tirucallane saponins,14,15 flavonoids, 12,16,17 phenolic,12 megastigmane glycosides,12,18,19 coumarins,20 and other compounds.21 These studies have highlighted the anti-inflammatory effects of P. scandens extracts on IL-12, p40, IL-6, and tumor necrosis factor (TNF)-α in LPS-stimulated bone marrow-derived dendritic macrophages (BMDCs)15 and the cytotoxic activity against human cancer macrophages lines, including KB (epidermoid carcinoma), SK-Mel-2 (melanoma), LU-1 (lung adenocarcinoma), and MCF7 (breast cancer).14
Based on previous primary studies showing good anti-inflammatory and immunological effects of the P. scandens extract and its isolated compounds, current study we continue to investigate bioactive compounds in this plant by choosing the highly potent extract of IL-1β and IL-10 cytokines activity in LPS-stimulated RAW 264.7 macrophages for isolation and purification, and we also investigate effects of the XL extract, its fractions, and isolated compounds on IL-1β and IL-10 cytokine-inducible production in RAW 264.7 macrophages.
Experimental
General experimental methods ‒ NMR spectra were recorded on Bruker AM 600 FT-NMR spectrometers (Karlsruhe, Germany) with TMS as an internal standard, J in Hz at 294 K. ESI-MS were analyzed with an LC-MS/MS and detector DAD (Shimadzu, Japan). Silica gel (Merck, 0.040‒0.063 mm particle size), RP-C18 (Merck, 30‒50 μm particle size), and MCI gel (Beijing Huideyi Technology Co., Ltd., CHP20P, 75‒150 μm) were used for column chromatography (CC). Thin-layer chromatography (TLC) was carried out with silica gel 60F254 (Merck, silica gel, 0.25 mm) and RP-18F254 (Merck, 0.25 mm) plates. Spots were detected under UV 254 and 366 nm or by spraying with 10% sulfuric acid in 96% ethanol (EtOH) and heating.
Plant materials ‒ The whole Paramignya scandens (Griff.) Craib (Rutaceae) plant was collected in Ma Da commune, Vinh Cuu district, Dong Nai province, Vietnam in December 2021. MSc. Nguyen Quynh Nga and MSc. Nguyen Van Hieu from the Medicinal Material Resources Center at NIMM taxonomically identified the plant. A voucher specimen (DV-091221) was deposited at the Herbarium of NIMM.
Extraction and bio-guided isolation ‒ The dried roots of P. scandens (3.0 kg) were extracted and refluxed three times with 70% EtOH for three hours (3 h) each time. The extracts were combined, filtered, and evaporated under reduced pressure, yielding a green residue (XLT, 290.0 g). This residue was suspended in water and partitioned with different organic solvents in ascending order of polarity, producing n-hexane (XLH, 10.0 g), ethyl acetate (XLE, 80.0 g), and aqueous (XLW, 120.0 g) extracts.
To investigate the effects of extracts (XLT, XLH, XLE, and XLW) on IL-1β and IL-10 cytokine production in RAW 264.7 macrophages, a preliminary macrophage proliferation assay was performed according to the protocol previously reported.21,22 The most active extract (XLE) was selected to be separated by a silica gel CC with a gradient mixture of n-hexane:acetone (20:1–1:1, v/v) of increasing polarity as eluents to give seven fractions (E1–E7). Fraction E2 (24.6 g) was fractionated by silica gel CC eluting with an isocratic mixture of n-hexane: acetone (15:1–1:1, v/v) to obtain five fractions (E2.1–E2.5). Fraction E2.2 (5.5 g) was applied to silica gel CC with an isocratic elution of n-hexane:ethyl acetate (15:1–1:1, v/v) to afford ten fractions (E2.2.1–E2.210) and compound 1 (184.0 mg). Compounds 2 (8.0 mg) and 7 (4.0 mg) were obtained from fraction E2.2.7 (56.0 mg) by using RP-C18 gel CC with an elution mixture of methanol: water (7:3, v/v). Fraction 2.2.4 (2.02 g) was applied to silica gel CC with an isocratic elution of n-hexane:ethyl acetate (4:1, v/v) to purify compound 6 (20.0mg). Fraction E2.3 (4.1 g) was applied to silica gel CC with an isocratic elution of n-hexane:ethyl acetate (15:1–1:1, v/v) to afford fourteen fractions (E2.3.1–E2.3.14). Purification of fraction 2.3.5 (354 mg) by RP-C18 gel CC with a mixture of methanol:water (7:3, v/v) to isolate two compounds 3 (12.0 mg) and 5 (9.0 mg). Fraction E2.4 (3.8 g) was applied to silica gel CC with an isocratic elution of n-hexane: acetone (10:1–1:1, v/v) to afford five fractions (E2.4.1–E2.4.5). Fraction E2.4.2 (486.0 mg) was fractionated by RP-C18 gel CC eluting with an isocratic mixture of methanol:water (15:1–1:1, v/v) to obtain compound 4 (84.0 mg).
Xanthyletin (1) ‒ Pale yellow solid; ESI-MS: 259.05 [M+H]+; 1H-NMR (600 MHz, CDCl3): δ 6.21 (1H, d, J = 9.6 Hz, H-3), 7.57 (1H, d, J = 9.6 Hz, H-4), 7.04 (1H, s, H-5), 6.71 (1H, s, H-8), 6.34 (1H, d, J = 10.2 Hz, H-1ʹ), 5.69 (1H, d, J = 9.6 Hz, H-2ʹ), 1.47 (6H, s, H-4ʹ, H-5ʹ); 13C-NMR (150 MHz, CDCl3): δ 161.2 (C-2), 131.2 (C-3), 143.3 (C-4), 124.8 (C-5), 118.5 (C-6), 156.9 (C-7), 120.8 (C-8), 155.5 (C-9), 113.0 (C-10), 112.7 (C-1ʹ), 104.4 (C-2ʹ), 77.4 (C-3ʹ), 28.4 (C-4ʹ, C-5ʹ).
Luvangetin (2) ‒ Pale yellow solid; ESI-MS: 259.05 [M+H]+; 1H-NMR (600 MHz, CDCl3): δ 6.23 (1H, d, J = 9.0 Hz, H-3), 7.56 (1H, d, J = 9.6 Hz, H-4), 6.83 (1H, s, H-5), 6.33 (1H, d, J = 10.2 Hz, H-1ʹ), 5.71 (1H, d, J = 9.6 Hz, H-2ʹ), 1.51 (6H, s, H-4ʹ, H-5ʹ), 3.98 (3H, s, 8-OCH3); 13C-NMR (150 MHz, CDCl3) δ: 160.6 (C-2), 113.0 (C-3), 143.6 (C-4), 119.1 (C-5), 119.1 (C-6), 149.3 (C-7), 135.7 (C-8), 148.3 (C-9), 113.2 (C-10), 121.1 (C-1ʹ), 131.2 (C-2ʹ), 77.8 (C-3ʹ), 28.2 (C-4ʹ, C-5ʹ), 61.4 (8-OCH3).
Clausenidin (3) ‒ Pale yellow solid; ESI-MS: 329.10 [M+H]+, 327.25 [M‒H]–; 1H-NMR (600 MHz, CDCl3): δ 6.16 (1H, d, J = 9.6 Hz, H-3), 8.04 (1H, d, J = 9.6 Hz, H-4), 2.75 (2H, s, H-2ʹ), 1.49 (6H, s, H-4ʹ, H-5ʹ), 4.90 (2H, dd, J = 18.6, 10.2 Hz, H-1ʺ), 6.22 (1H, dd, J = 17.4, 10.8 Hz, H-2ʺ), 1.64 (6H, s, H-4ʺ, H-5ʺ), 12.97 (1H, s, OH); 13C-NMR (150 MHz, CDCl3): δ 160.7 (C-2), 110.8 (C-3), 138.6 (C-4), 159.1 (C-5), 104.1 (C-6), 160.0 (C-7), 114.6 (C-8), 160.0 (C-9), 103.3 (C-10), 198.3 (C-1ʹ), 47.7 (C-2ʹ), 80.0 (C-3ʹ), 26.5 (C-4ʹ, C-5ʹ), 108.4 (C-1ʺ), 149.6 (C-2ʺ), 41.0 (C-3ʺ), 29.5 (C-4ʺ, C-5ʺ).
Nordentatin (4) ‒ Pale yellow solid; ESI-MS: 327.15 [M+H]+, 325.30 [M‒H]–; 1H-NMR (600 MHz, CDCl3): δ 6.14 (1H, d, J = 9.6 Hz, H-3), 8.01 (1H, d, J = 9.6 Hz, H-4), 6.50 (1H, d, J = 10.2 Hz, H-1ʹ), 5.69 (1H, d, J = 10.2 Hz, H-2ʹ), 1,44 (6H, s, H-4ʹ, H-5ʹ), 4.85 (1H, d, J = 11.4 Hz, H-1ʺa), 4.92 (1H, d, J = 18.6 Hz, H-1ʺb), 6.28 (1H, dd, J = 17.4, 10.8 Hz, H-2ʺ), 1.64 (6H, s, H-4ʺ, H-5ʺ); 13C-NMR (150 MHz, CDCl3): δ 161.4 (C-2), 110.3 (C-3), 139.2 (C-4), 146.6 (C-5), 106.1 (C-6), 155.9 (C-7), 116.2 (C-8), 154.2 (C-9), 103.9 (C-10), 115.0 (C-1ʹ), 130.0 (C-2ʹ), 77.2 (C-3ʹ), 27.3 (C-4ʹ, C-5ʹ), 108.1 (C-1ʺ), 150.1 (C-2ʺ), 41.0 (C-3ʺ), 29.6 (C-4ʺ, C-5ʺ).
Dentatin (5) ‒ Pale yellow solid; ESI-MS: 313.10 [M+H]+, 311.15 [M‒H]–; 1H-NMR (600 MHz, CDCl3): δ 6.19 (1H, d, J = 9.6 Hz, H-3), 7.87 (1H, d, J = 9.6 Hz, H-4), 6.57 (1H, d, J = 9.6 Hz, H-1ʹ), 5.69 (1H, d, J = 10.2 Hz, H-2ʹ), 1.40 (6H, s, H-4ʹ, H-5ʹ), 4.88 (1H, d, J = 12.0 Hz, H-1ʺa), 4.94 (1H, d, J = 18.6 Hz, H-1ʺb), 6.30 (1H, dd, J = 17.4, 10.8 Hz, H-2ʺ), 1.66 (6H, s, H-4ʺ, H-5ʺ), 3.83 (3H, s, 5-OCH3); 13C-NMR (150 MHz, CDCl3): δ 160.7 (C-2), 111.7 (C-3), 138.9 (C-4), 151.2 (C-5), 108.2 (C-6), 156.0 (C-7), 119.2 (C-8), 154.0 (C-9), 107.6 (C-10), 116.4 (C-1ʹ), 130.4 (C-2ʹ), 77.4 (C-3ʹ), 27.6 (C-4ʹ, C-5ʹ), 108.2 (C-1ʺ), 149.9 (C-2ʺ), 41.2 (C-3ʺ), 29.5 (C-4ʺ, C-5ʺ), 63.4 (5-OCH3).
Clausarin (6) ‒ Pale yellow solid; ESI-MS: 381.15 [M+H]+, 379.25 [M‒H]–; 1H-NMR (600 MHz, CDCl3): δ 7.84 (1H, s, H-4), 5.07 (1H, dd, J = 10.2; 1.2 Hz, H-11a), 5.11 (1H, dd, J = 17.4, 1.2 Hz, H-11b), 6.18 (1H, dd, J = 17.4, 10.8 Hz, H-12) 1.42 (6H, s, H-14, H-15), 6.51 (1H, d, J = 9.6 Hz, H-1ʹ), 5.66 (1H, d, J = 9.6 Hz, H-2ʹ), 1.47 (6H, s, H-4ʹ, H-5ʹ), 4.85 (1H, dd, J = 10.8, 1.2 Hz, H-1ʺa), 4.93 (1H, dd, J = 17.4, 1.2 Hz, H-1ʺb), 6.29 (1H, dd, J = 17.4, 10.8 Hz, H-2ʺ), 1.64 (6H, s, H-4ʺ, H-5ʺ); 13C-NMR (150 MHz, CDCl3): δ 159.8 (C-2), 129.0 (C-3), 132.9 (C-4), 146.5 (C-5), 105.9 (C-6), 154.8 (C-7), 115.3 (C-8), 153.4 (C-9), 104.0 (C-10), 111.9 (C-11), 145.7 (C-12), 40.3 (C-13), 26.1 (C-14, C-15), 115.5 (C-1ʹ), 129.7 (C-2ʹ), 77.0 (C-3ʹ), 27.3 (C-4ʹ, C-5ʹ), 107.9 (C-1ʺ), 150.2 (C-2ʺ), 40.9 (C-3ʺ), 29.5 (C-4ʺ, C-5ʺ).
Anisocoumarin E (7) ‒ Pale yellow powder; 1H-NMR (600 MHz, CDCl3): δ 6.21 (1H, d, J = 9.6 Hz, H-3), 7.62 (1H, d, J = 9.6 Hz, H-4), 7.34 (1H, d, J = 8.4 Hz, H-5), 6.83 (1H, d, J = 8.4 Hz, H-6), 4.57 (2H, d, J = 7.2 Hz, H-1ʹ), 5.40 (1H, t, J = 6.6 Hz, H-2ʹ), 1.77 (3H, s, H-4ʹ), 1.71 (3H, s, H-5ʹ), 4.00 (2H, s, H-1ʺ), 2.80 (1H, t, J = 7.2 Hz, H-3ʺ), 1.19 (6H, d, J = 6.6 Hz, H-4ʺ, H-5ʺ); 13C-NMR (150 MHz, CDCl3): δ 161.0 (C-2), 112.8 (C-3), 143.7 (C-4), 127.3 (C-5), 108.4 (C-6), 159.9 (C-7), 112.9 (C-8), 153.4 (C-9), 112.3 (C-10), 65.7 (C-1ʹ), 119.0 (C-2ʹ), 138.6 (C-3ʹ), 18.4 (C-4ʹ), 25.7 (C-5ʹ), 34.8 (C-1ʺ), 210.7 (C-2ʺ), 40.9 (C-3ʺ), 18.2 (C-4ʺ, C-5ʺ).
Cell culture ‒ RAW 264.7 macrophages (ATCC, Manassas, VA, USA) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Sigma Aldrich, St. Louis, MO, USA), 10% fetal bovine serum (FBS, Sigma Aldrich, St. Louis, MO, USA), penicillin G (100 U/mL), streptomycin (100 μg/mL) in an incubator stable at 37°C, 5% CO2.
Assay for IL-1β and IL-10 cytokine-inducible activity in RAW 264.7 macrophages ‒ The RAW 264.7 macrophages were seeded in 96-well tissue culture plates at a density of 104 macrophages/well with 10% FBS and incubated at 37°C and 5% CO2 for 12 h. Subsequently, the macrophages were stimulated with lipopolysaccharide (LPS, S igma Aldrich, St. Louis, MO, USA) 5 ng/mL in the presence or absence of the extracts at a concentration of 20 μg/mL or the isolated compounds at different concentrations. After incubating for 24 h, the culture supernatant was collected, and the levels of IL-1β and IL-10 cytokines were quantified using a commercially available ELISA kit following the manufacturer’s instructions.21,22 RAW 264.7 macrophages incubated with DMSO 1% (Sigma Aldrich, St. Louis, MO, USA) and LPS 5 ng/mL were used as a control group. The assay was performed in quintuplicate and independently analyzed.21,22
Statistical analysis ‒ Quantitative data are presented as the mean ± SEM and analyzed based on the one-way analysis of variance (ANOVA) and followed by Dunnett’s test using GraphPad Prism 9.5.0. Statistical significance was defined as p values less than 0.05.
Results and Discussion
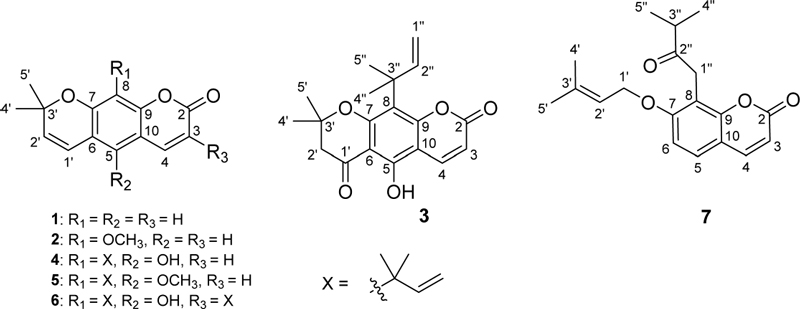
The results showed that the XLE extract exhibited stronger inhibition of LPS-induced IL-1β production in RAW 264.7 macrophages compared to the other extracts, as depicted in Fig. 1A. The order of inhibitory activity was found to be XLE (78.97%) ‒ XLH (79.97%) > XLW (77.06%) > XLT (74.18%) at a concentration of 20 μg/mL. Moreover, the XLE extract also showed stronger stimulation of LPS-induced IL-10 production in RAW 264.7 macrophages compared to the other extracts, as shown in Fig. 1B. Specifically, the order of stimulatory activity was exhibited to be XLE (71.66%) ‒ XLW (70.78%) > XLH (66.97%) > XLT (59.60%) at a concentration of 20 μg/mL. Based on these findings, the XLE extract was selected for further isolation of compounds by using silica gel and RP-C18 gel column chromatography.
Modulatory effects of the P. scandens ethanol extract and its fractions on LPS-induced IL-1β and IL-10 cytokine production in RAW 264.7 macrophages. Ethanol extract (XLT); n-hexane fraction (XLH), ethyl acetate fraction (XLE), and water fraction (XLW). Values represent the mean ± SEM (n = 5). Statistical significance was analyzed by one-way ANOVA followed by the Turkey test. ns: not statistical significance and ***p < 0.001 compared to the cells treated with LPS only.
From the most activity EtOAc extract (XLE) on IL-1β and IL-10 cytokine production, seven coumarins were isolated by using various silica gel and RP-C18 gel chromatographic columns (Fig. 2). The chemical structures were verified based on MS and NMR experimented spectra data and comparison to those of reported data.
compound 1 was isolated as a pale yellow powder. The ESI-MS spectrum showed a quasi-molecular peak at m/z 259.05 [M+H]+, and when combined with NMR data, it suggested that the molecular formula of 1 was C14H12O3. The 1H-NMR spectrum of 1 displayed signals characteristic of the 1-benzopyran-2-one skeleton, which was a defining feature of the coumarin group. These signals included two cis-olefinic protons at δH 6.21 (1H, d, J = 9.6 Hz, H-3) and 7.57 (1H, d, J= 9.6 Hz, H-4), as well as two singlet olefinic protons at δH 7.04 (1H, s, H-5) and 6.71 (1H, s, H-8). Additionally, signals corresponding to the methine groups of the 3,6-dihydro-2H-pyran ring were observed at δH 6.34 (1H, d, J = 10.2 Hz, H-1ʹ) and 5.69 (1H, d, J = 9.6 Hz, H-2ʹ), along with two methyl groups at δH 1.47 (6H, s, H-4ʹ, H-5ʹ). The 13C-NMR spectrum displayed signals of 14 carbons, including four signals characteristic of 1-benzopyran-2-one [δC 131.2 (C- 3), 143.3 (C-4), 124.8 (C-5), and 120.8 (C-8)], and five signals corresponding to aromatic carbons at δC 161.2 (C-2), 118.5 (C-6), 156.9 (C-7), 155.5 (C-9), and 113.0 (C-10). The carbons of the -CH and -C groups of the 3,6-dihydro-2H-pyran ring were observed at δC 112.7 (C-1ʹ), 104.4 (C-2ʹ), and 77.4 (C-3ʹ). The signals at δC 28.4 (C-4ʹ, C-5ʹ) were specific to the methyl groups. Consequently, the structure of 1 was identified as xanthyletin based on spectral analysis and comparison with literature reports.23
compound 2 was obtained as a pale yellow powder and had the molecular formula C15H14O4, which was confirmed through ESI-MS analysis, revealing a quasi-molecular ion peak at m/z 259.05 [M+H]+. The NMR spectra of 2 resembled those of 1, except that 2 exhibited a singlet olefinic proton. Moreover, 2 displayed a signal corresponding to the methoxy group at δH 3.98 and δC 61.4. The position of the methoxy group was determined to be at C-8 based on the HMBC correlation observed between the proton at δH 3.98 and C-8 (δC 135.7). Therefore, the structure of 2 was identified as luvangetin using spectral analysis and in comparison with previously reported literature.24
compound 3 was isolated as pale yellow powder. Its ESI-MS spectrum displayed quasi-molecular ion peaks at m/z 329.10 [M+H]+ and 327.25 [M‒H]–. These, combined with NMR data, suggested that the molecular formula of 3 was C19H20O5. The 1H-NMR spectra of 3 exhibited signals corresponding to the 1-benzopyran-2-one skeleton, like 1 and 2. Additionally, 3 displayed signals of a CH2 group of the 2,3-dihydro-4H-pyran-4-one ring at δH 2.75 (2H, s) and two methyl groups at δH 1.49 (6H, s, H-4ʹ, H-5ʹ). Signals at δH 4.90 (2H, dd, J = 18.6, 10.2 Hz, H-1ʺ), 6.22 (1H, dd, J = 17.4, 10.8 Hz, H-2ʺ), and 1.64 (6H, s, H-4ʺ, H-5ʺ) were specific to the 1,1-dimethylallyl group. The signal at δH 12.97 (1H, s, OH) indicated the presence of a hydroxyl group. The 13C-NMR spectrum exhibited signals of 19 carbons, including 9 signals of the 1-benzopyran-2-one skeleton and 7 aromatic carbons [δC 160.7 (C-2), 159.1 (C-5), 104.1 (C-6), 160.0 (C-7), 114.6 (C-8), 160.0 (C-9), and 103.3 (C-10)]). Carbons of the -CH2 and -C groups of the 2,3-dihydro-4H-pyran-4-one ring were observed at δC 198.3 (C-1ʹ), 47.7 (C-2ʹ), and 80.0 (C-3ʹ). The two methyl groups (δC 26.5) of the 2,3-dihydro-4Hpyran-4-one ring at C-3ʹ were determined through the HMBC correlations between the methyl proton (δH 1.49) and the carbons at δC 80.0 (C-3ʹ) and 47.7 (C-2ʹ). Signals of the 1,1-dimethylallyl group were also observed at δC 108.4 (C-1ʺ), 149.6 (C-2ʺ), 41.0 (C-3ʺ), and 29.5 (C-4ʺ, C-5ʺ). The position of this group was determined to be at C-8 through the HMBC interactions between H-2ʺ (δH 6.22)/H-4ʺ (δH 1.64), and C-8 (δC 114.6). Comparing the NMR spectra,25 the structure of 3 was identified as clausenidin.
compound 4 was obtained as a pale yellow powder. Its molecular formula was deduced to be C19H20O4 based on the presence of pseudo-molecular ion peaks at m/z 313.10 [M+H]+ and 311.15 [M‒H]– in conjunction with NMR spectra. The 1H-NMR spectra of 4 exhibited signals corresponding to the 1-benzopyran-2-one skeleton, two methine groups of the 3,6-dihydro-2H-pyran ring, and two methyl groups, which were analogous to those in 1. Moreover, 4 also featured signals of the 1,1-dimethylallyl group attached at C-8, similar to 3. The 13C-NMR spectrum of 4 displayed signals from 19 carbons, similar to 3. However, in comparison with 3, 4 exhibited a slight alteration in the structure of the 3,6-dihydro-2H-pyran ring. Based on the analysis outlined above and by comparing the NMR spectra,26 the structure of 4 was identified as nordentatin.
compound 5 was isolated as a pale yellow powder. Its molecular formula was determined to be C20H22O4 based on the presence of quasi-molecular ion peaks at m/z 327.15 [M+H]+ and 325.30 [M‒H]–, as well as NMR spectra. The NMR spectrum of 5 closely resembled that of 4; however, 5 exhibited a signal for the methoxy group instead of the hydroxyl group at δH 3.83 (3H, s) and δC 63.4. Additionally, 5 had only two methylene groups instead of the four groups present in the structure of 4. The ESI-MS spectrum also displayed a mass difference of 14 units, further confirming the substitution of the -OH group with the -OCH3 group in the structure of 5. Therefore, 5 was identified as dentatin by comparing its NMR data with published data.27
compound 6 was obtained as a pale yellow powder. Its molecular formula, C24H28O4, was determined based on NMR data and the presence of pseudo-molecular ion peaks at m/z 381.15 [M+H]+ and 379.25 [M‒H]–. The NMR spectra of 6 exhibited similarities to those of 4. However, the NMR spectrum of 6 revealed the presence of a proton signal from the 1-benzopyran-2-one skeleton. Furthermore, 6 displayed signals of another 1,1-dimethylallyl group at δH 5.07 (1H, dd, J = 10.2, 1.2 Hz, H-11), 5.11 (1H, dd, J = 17.4, 1.2 Hz, H-11), 6.18 (1H, dd, J = 17.4, 10.8 Hz, H-12), 1.42 (6H, s, H-14, H-15), and δC 111.9 (C-11), 145.7 (C-12), 40.3 (C-13), and 26.1 (C-14, C-15). The position of this group was determined to be at C-3, confirmed by HMBC interactions between H-12 (δH 6.18) and C-3 (δC 129.0), as well as between H-4 (δH 7.84) and C-13 (δC 40.3). Thus, 6 was identified as clausarin by comparing its NMR data with published data.28
compound 7 was isolated as a pale yellow powder. The 1H-NMR spectra of 7 revealed signals of the 1-benzopyran-2-one skeleton, which was characteristic of the coumarin group. Additionally, 7 displayed signals of the prenyloxy (3-methylbut-2-enyloxy) group [δH 4.57 (2H, d, J = 7.2 Hz, H-1ʹ), 5.40 (1H, t, J = 6.6 Hz, H-2ʹ), 1.77 (3H, s, H-4ʹ), and 1.71 (3H, s, H-5ʹ)], as well as the 3-methyl-2-oxobutyl group [δH 4.0 (2H, s, H-1ʺ), 2.80 (1H, t, J = 7.2 Hz, H-2ʺ), and 1.19 (6H, d, J = 6.6 Hz, H-4ʺ, H-5ʺ)]. The 13C-NMR spectrum of 7 exhibited signals of 19 carbons, including 9 carbons of the 1-benzopyran-2-one skeleton. Signals of the prenyloxy group were observed at δC 65.7 (C-1ʹ), 119.0 (C-2ʹ), 138.6 (C-3ʹ), 18.4 (C-4ʹ), and 25.7 (C-5ʹ). The position of this group was determined to be at C-7 through HMBC interactions between H-1ʹ (δH 4.57) / H-5 (δH 7.34) and C-7 (δC 159.9). Furthermore, the position of the 3-methyl-2-oxobutyl group [δC 34.8 (C-1ʺ), 210.7 (C-2ʺ), 40.9 (C-3ʺ), and 18.2 (C-4ʺ, C-5ʺ)] was determined to be at C-8 through HMBC correlations between H-1ʺ (δH 4.00) and C-7 (δC 159.9)/C-8 (δC 112.9)/C-9 (δC 153.4). Based on spectral analysis and by comparison with published literature,29 7 was identified as anisocoumarin E.
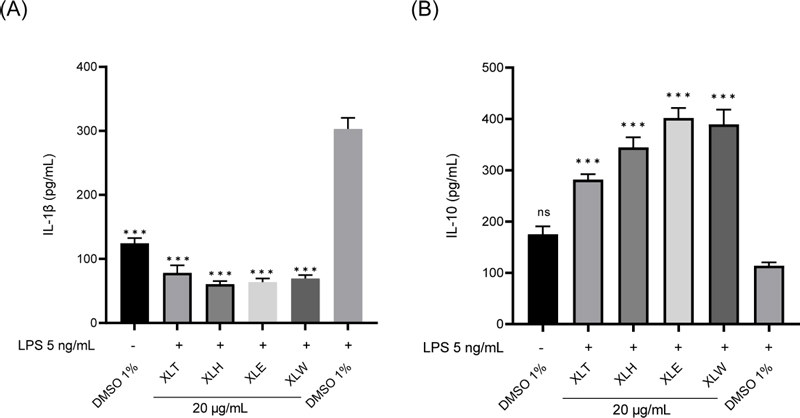
Seven coumarins (1–7) were isolated from XLE and evaluated for their effects on IL-1β and IL-10 cytokine production in RAW 264.7 macrophages. The results were presented in Fig. 3A, compounds 1–7 at the concentration of 10 μM exhibited significant inhibitory activity on IL-1β production in RAW 264.7 macrophages, with IL-1β concentrations ranging from 42.77 to 69.76 pg/mL compared to the control group treated with LPS 5 ng/mL (IL-1β concentration of 344.85 pg/mL). Among these compounds, compounds 1, 2, and 6 demonstrated stronger inhibition of IL-1β secretion than others, with an IL-1β concentration of 58.05 pg/mL, 42.77 pg/mL, and 53.83 pg/mL, respectively. Furthermore, the effect of the seven compounds (1–7) on the stimulation of LPS-induced IL-10 production in RAW 264.7 macrophages was assessed and illustrated in Fig. 3B. Again, all compounds at the concentration of 10 μM significantly stimulated IL-10 secretion in LPS-induced RAW 264.7 macrophages, with IL-10 levels ranging from 175.98 to 321.56 pg/mL, compared to the control group treated with LPS alone (IL-10 concentration of 84.70 pg/mL). Notably, coumarins 1, 2, and 6 also exhibited stronger IL-10 stimulation than others, with an IL-10 concentration of 226.26 pg/mL, 321.56 pg/mL, and 291.28 pg/mL, respectively.
Modulatory effects of the isolated compounds (1–7) on LPS-induced IL-1β and IL-10 cytokine production in RAW 264.7 macrophages. Values represent the mean ± SEM (n = 5). Statistical significance was analyzed by one-way ANOVA followed by the Turkey test. ns: not statistical significance and ***p < 0.001 compared to the cells treated with LPS only.
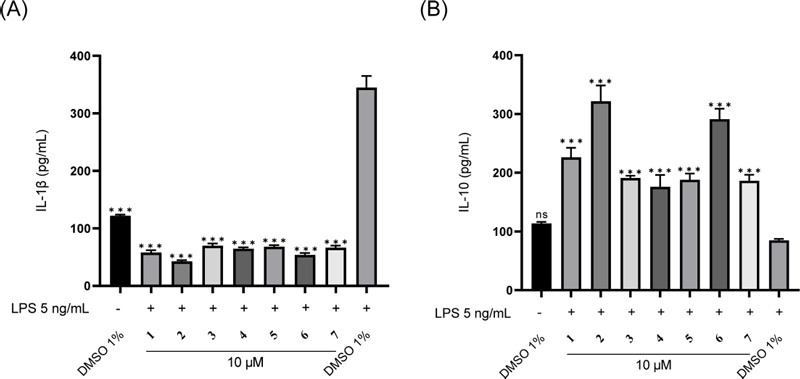
The evaluation results revealed that compounds 1, 2, and 6 exhibited potential stimulation of IL-10 secretion (increased by > 2.5 times) and inhibition of IL-1β secretion (reduced by > 5.9 times) compared to the LPS control group. Therefore, further evaluation of these compounds was conducted on cytokine IL-10 and IL-1β at different concentrations (1–30 μM). The results (Fig. 4) showed that the stimulatory effect on IL-10 production and inhibitory effect on IL-1β production of these compounds gradually increased with the test concentration in both RAW 264.7 macrophages and LPS-induced RAW 264.7 macrophages. Compounds 1, 2, and 6 inhibited IL-1β production in LPS-induced RAW 264.7 macrophages with IC50 values of 10.70 ± 1.18 μM, 8.57 ± 1.05 μM, and 17.43 ± 1.05 μM, respectively.
Modulation effects of compounds (1, 2, and 6) on LPS-induced IL-1β and IL-10 production in RAW 264.7 macrophages.(A) Stimulatory effect of compounds on LPS-induced IL-10 cytokine production in RAW 264.7 macrophages. RAW 264.7 cells were stimulated by LPS (5 ng/mL) in the absence (DMSO) or presence of compounds (1, 2, and 6 at 1–30 μM) for 12 h. Values represent the mean ± SEM (n = 5). Statistical significance was analyzed by one-way ANOVA followed by the Turkey test. *p < 0.05; **p < 0.01; ***p < 0.001, treatment + LPS vs LPS, +p < 0.05; ++p < 0.01; +++p < 0.001, treatment vs DMSO.(B) Inhibitory effect of compounds on LPS-induced IL-1β cytokine production in RAW 264.7 macrophages. RAW 264.7 cells were separately treated with DMSO or LPS (5 ng/mL) or compounds (1, 2, and 6 at 1–30 μM) for 12 h. Values represent the mean ± SEM (n = 5). Statistical significance was analyzed by one-way ANOVA followed by the Turkey test. ###p < 0.001 LPS vs DMSO, *p < 0.05; **p < 0.01; ***p < 0.001, treatment + LPS vs LPS, +p < 0.05; ++p < 0.01; +++p < 0.001, treatment vs DMSO.
In comparison to previous studies, that have identified some of these compounds, such as compounds 3, 6, and 7, in the Clausena genus,25,29,30 in this study, we focused on the chemical analysis based on the biological activity of P. scandens roots, leading to the isolation and identification of seven coumarins (1–7). Among these compounds, three compounds (3, 6, and 7) were isolated for the first time from the Paramignya genus, and two coumarins (1 and 2) were reported for the first time from this species. Besides, previous studies have highlighted the effects of P. scandens extracts on IL-12, p40, IL-6, and TNF-α in LPS15 and the cytotoxic activity against human cancer macrophage lines.14 Likewise, the results in this study also demonstrated the potential of all the compounds (1–7), as well as the roots of P. scandens, in IL-1β and IL-10 cytokine-inducible activity.
Besides, in previous studies, the seven isolated compounds have a characteristic coumarin structure in the Paramygnya genus and this is also the main component of this genus. This group of substances also plays an important role related to the biological effects of species of the genus Paramygnya.14,15 Besides, the results in this study show that the compounds also showed significant effects on LPS-induced IL-1β and IL-10 cytokine production in RAW 264.7 macrophages, suggesting that the characteristic 1-benzopyran-2-one skeleton of the coumarin group may play an important role in the effects on LPS-induced IL-1β and IL-10 cytokine production and is the structural part that provides the effect (pharmacophore) of compounds in this group.
In conclusion, seven coumarin compounds were isolated from P. scandens. All isolates (1‒7) showed significant effects on modulation of LPS-induced IL-1β and IL-10 cytokines production in RAW 264.7 macrophages. The results contributed to expanding the knowledge of the chemistry and bioactivities of P. scandens and provided valuable data for future investigations on this species.
Acknowledgments
This study was financially supported by the Ministry of Science and Technology Vietnam [NĐT.85.KR/20, contract no. 09/2020/HĐ-NĐT signed 09/9/2020-MOST], and The National Research Foundation of Korea (NRF) grant funded by the Korean government [2019K1A3A1A05088041].
Conflict of interest
The authors declare that they have no conflicts of interest.
References
- Abbas, A. K.; Lichtman, A. H.; Pillai, S. Cellular and Molecular Immunology e-book; Elsevier Health Sciences; Philadelphia, Pennsylvania, 2021; 26–130.
-
Lopez-Castejon, G.; Brough, D. Cytokine Growth Factor Rev. 2011, 22, 189–195.
[https://doi.org/10.1016/j.cytogfr.2011.10.001]

-
Ben-Sasson, S. Z.; Hu-Li, J.; Quiel, J.; Cauchetaux, S.; Ratner, M.; Shapira, I.; Dinarello, C. A.; Paul, W. E. Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 7119–7124.
[https://doi.org/10.1073/pnas.0902745106]

-
Galozzi, P.; Bindoli, S.; Doria, A.; Sfriso, P. Autoimmun. Rev. 2021, 20, 102785.
[https://doi.org/10.1016/j.autrev.2021.102785]

-
Sabat, R.; Grutz, G.; Warszawska, K.; Kirsch, S.; Witte, E.; Wolk, K.; Geginat, J. Cytokine Growth Factor Rev. 2010, 21, 331−344.
[https://doi.org/10.1016/j.cytogfr.2010.09.002]

-
Eskdale, J.; Kube, D.; Tesch, H.; Gallagher, G. Immunogenetics 1997, 46, 120−128.
[https://doi.org/10.1007/s002510050250]

-
Abrams, J., Figdor, C. G., De Waal Malefyt, R., Bennett, B., De Vries, J. E., J. Exp. Med. 1991, 174, 1209–1220.
[https://doi.org/10.1084/jem.174.5.1209]

- Jenkins, J. K.; Malyak, M.; Arend, W. P. Lymphokine Cytokine Res. 1994, 13, 47−54.
-
Sun, Y.; Ma, J.; Li, D.; Li, P.; Zhou, X.; Li, Y.; He, Z.; Qin, L.; Liang, L.; X, Luo, J. Neuroinflammation 2019, 16, 1–13.
[https://doi.org/10.1186/s12974-019-1452-1]

-
Gao, Y.; Tu, D.; Yang, R.; Chu, C.-H.; Hong, J.-S.; Gao, H.-M. Int. J. Mol. Sci. 2020, 21, 465.
[https://doi.org/10.3390/ijms21020465]

-
Akdis, C. A.; Blaser, K. Immunology 2001, 103, 131−136.
[https://doi.org/10.1046/j.1365-2567.2001.01235.x]

- Nguyen, H. T. P.; Nguyen, T. D. T.; Nong, V. D.; Ninh, T. N.; Nguyen, V. D.; Nguyen, X. C.; Nguyen, H. N.; Phan, V. K.; Chau, V. M. J. Chem. 2015, 53, 84−89.
- Nguyen, T. D. T. Research on the Chemical Composition and Biological Activities of Paramignya scandens (Griff.) Craib) in Lam Dong, Doctoral thesis, Institute of Marine Biochemistry, 2015, 8–27.
- Nguyen, H. T. P.; Nguyen, T. D. T.; Ninh, T. N.; Pham, T. M. H.; Nguyen, P. T.; Nguyen, X. C.; Nguyen, V. T.; Nguyen, H. N.; Phan, V. K.; Chau, V. M. Phytochem. Lett. 2014, 9, 78−81.
- Nguyen, H. T. P.; Nguyen, T. D. T.; Ninh, T. N.; Nguyen, P. T.; Kim, S.; Koh, Y. S.; Nguyen, V. T.; Nguyen, X. C.; Nguyen, H. N.; Phan, V. K. Chem. Pharm. Bull. 2015, 63, 558−564.
-
Bowen, I. H.; Patel, Y. N. J. Pharm. Pharmacol. 1998, 50, 232−232.
[https://doi.org/10.1111/j.2042-7158.1998.tb02432.x]

-
Wattanapiromsakul, C.; Waterman, P. G. Phytochemistry 2000, 55, 269−273.
[https://doi.org/10.1016/S0031-9422(00)00311-3]

-
Kumar, V.; Niyaz, N. M.; Saminathan, S.; Wickramaratne, D. B. Phytochemistry 1998, 49, 215−218.
[https://doi.org/10.1016/S0031-9422(97)00760-7]

-
Kumar, V.; Niyaz, N. M.; Wickramaratne, D. B. Phytochemistry 1995, 38, 805−806.
[https://doi.org/10.1016/0031-9422(94)00485-C]

- Trinh, T. D.; Luong, V. D.; Huynh, T. T. Zhong Yao Cai 2016, 1+2, 35–40.
-
Jin, Q.; Lee, C.; Lee, J. W.; Yeon, E. T.; Lee, D.; Han, S. B.; Hong, J. T.; Kim, Y.; Lee, M. K.; Hwang, B. Y. J. Nat. Prod. 2014, 77, 1724−1728.
[https://doi.org/10.1021/np400831p]

-
Jeon, Y. J.; Han, S. H.; Lee, Y. W.; Lee, M.; Yang, K. H.; Kim, H. M. Immunopharmacology 2000, 48, 173−183.
[https://doi.org/10.1016/S0162-3109(00)00199-5]

- Tran, T. T. Q.; Le, T. K. T.; Pham, D. P. Pharm. J. 2014, 54, 60−65.
- Hoang, L. T. A.; Kim, D.; Ko, W.; Tran, M. H.; Nguyen, X. N.; Pham, H. Y.; Bui, H. T.; Luu, H. T.; Vu, N. L.; Tran, G. Pharm. Biol. 2017, 55, 1195−1201.
-
Lim, P. C.; Ramli, H.; Kassim, N. K.; Ali, Z.; Khan, I. A.; Shaari, K.; Ismail, A. Biochem. Syst. Ecol. 2019, 82, 52−55.
[https://doi.org/10.1016/j.bse.2018.12.010]

-
Promsuwan, P.; Yenjai, C. Asian J. Chem. 2013, 25, 3629.
[https://doi.org/10.14233/ajchem.2013.13687]

-
Songsiang, U.; Thongthoom, T.; Zeekpudsa, P.; Kukongviriyapan, V.; Boonyarat, C.; Wangboonskul, J.; Yenjai, C. ScienceAsia 2012, 38, 75−81.
[https://doi.org/10.2306/scienceasia1513-1874.2012.38.075]

-
Takemura, Y.; Kawaguchi, H.; Maki, S.; Ju-ichi, M.; Omura, M.; Ito, C.; Furukawa, H. Chem. Pharm. Bull. 1996, 44, 804−809.
[https://doi.org/10.1248/cpb.44.804]

-
Ngadjui, B. T.; Ayafor, J. F.; Sondengam, B. L.; Connolly, J. D. J. Nat. Prod. 1989, 52, 243−247.
[https://doi.org/10.1021/np50062a003]

-
Su, C.-R.; Yeh, S. F.; Liu, C. M.; Damu, A. G.; Kuo, T.-H.; Chiang, P. C.; Bastow, K. F.; Lee, K. H.; Wu, T. S.; Bioorg. Med. Chem. 200817, 6137–6143.
[https://doi.org/10.1016/j.bmc.2008.12.007]