Development and Validation of an HPLC-DAD Method for Simultaneous Quantitation of Steppogenin and Flavonoids from the Stems of Morus alba
Abstract
Morus alba L. is well-known for its medicinal and economic value, particularly in Asian countries. Among the isolated compounds from this plant, steppogenin is exhibited as a flavonoid with promising pharmacological properties. This study focused on isolating bioactive compounds, notably steppogenin, from the ethyl acetate extract of M. alba. Additionally, a high-performance liquid chromatography-diode array detector (HPLC-DAD) method for the simultaneous quantification of steppogenin and isolated compounds was developed and validated. The calibration curve showed excellent linearity, with a correlation coefficient (R2) value greater than 0.9957. The limit of detection (LOD) ranged from 0.006 to 0.018 μg/mL, whereas the limit of quantification (LOQ) ranged from 0.020 to 0.061 μg/mL. In precision tests conducted intra-day and inter-day, the accuracy was between 97.32% and 106.39%, with relative standard deviations (RSD) less than 2.27% and 1.65%, respectively. The presence of steppogenin and other flavonoids was confirmed by the study, contributing to the understanding of the chemical composition of M. alba. This validated analytical method offers a reliable means of quantifying steppogenin and aiding future research into its therapeutic potential.
Keywords:
Morus alba, Steppogenin, Flavonoids, HPLC-DADIntroduction
Morus alba L., commonly known as mulberry, belongs to the Moraceae family and is extensively distributed throughout Asia, primarily in Japan, China, Korea, and Vietnam.1 In China, M. alba holds significant economic importance. Its leaves serve as a vital food source for domesticated silkworms, while its root bark is found in multiple applications in traditional Chinese medicine, where it is used for treating conditions such as cancer, neurological diseases, and diabetes.2,3 In Korea, M. alba has been an ingredient in traditional medicine for the treatment of cough, asthma, hematemesis, dropsy, and hypertension.4 Previous studies on the phytochemical composition of M. alba have led to the isolation of various polyphenolic compounds, including flavonoids, benzofurans, and Diels-Alder type adducts.2–5 These compounds exhibit diverse pharmacological activities such as anti-diabetic, antioxidant, anti-inflammatory, and immunological enhancement properties.
Flavonoids, the main constituents of this plant, have been reported to exhibit various biological effects. Steppogenin, a unique flavonoid found in different parts of M. alba, has been demonstrated to have promissing inhibitory activity against tyrosinase and α-glucosidase.6,7 Additionally, steppogenin has been shown to suppress the transcriptional activity of hypoxia-inducible factor-1α (HIF-1α) under hypoxic conditions in HEK293T cells and inhibit vascular endothelial growth factor (VEGF)-induced DLL4 expression in vascular endothelial cells (ECs) in a dose-dependent manner.8
In a previous study, Zhou et al. reported a comprehensive analysis of the chemical composition from different parts of M. alba, including roots, leaves, twigs, and fruits.9 They employed high-performance liquid chromatography-diode array detectors (HPLC-DAD) coupled with the quadrupole time-of-flight mass spectrometry (Q-TOF-MS) to examine these constituents. Their findings revealed the presence of 60 secondary metabolites, some of which showed promising hypoglycemic activities. However, the quantity of steppogenin was not determined.
In our present study, we aimed to extract and isolate compounds from the ethyl acetate (EtOAc) extract of M. alba, which contains the highest amount of steppogenin. Moreover, an HPLC-DAD method was developed to simultaneously quantify steppogenin and the isolated compounds. Validation tests, including linearity, limits of detection and quantification, precision, and accuracy, were also performed to emphasize the reproducibility and precision of our developed method.
Experimental
General experimental procedures – Experiments in this study were carried out in accordance with the procedures we previously detailed.10,11 1H- and 13C- nuclear magnetic resonance (NMR) measurements were conducted using a Bruker Avance Digital 500 MHz spectrometer (Karlsruhe, Germany), using TMS as an internal standard. The isolation of compounds was achieved through column chromatography (CC) utilizing silica gel 60 (0.063–0.200 mm) and LiChroprep RP-18 (40–63 μm) (Merck, Darmstadt, Germany). Thin-layer chromatography was carried out on TLC glass plates coated with silica gel 60 F254 and RP-18 F254S (Merck). Spot detection was performed under UV light at 254 and 356 nm, and visualization was accomplished by spraying the plates with 10% H2SO4 and heating at 110°C for 1–1.5 minutes. Chemicals and solvents used in CC experiments were acquired from Daejung Chemicals (Gyeonggi-do, Korea). HPLC grade methanol was purchased from Burdick & Jackson (Muskegon, MI, USA). The water used in the study was purified with a Milli-Q water purification system from Merck Millipore (Merck).
Plant material – Dried stems of M. alba were purchased from the Bonchowon herbal medicine store (Korea). The plant material was authenticated by Professor Byung Sun Min, Daegu Catholic University. The voucher specimen (accession code: 23B-MA) was deposited at the Pharmacognosy Laboratory, College of Pharmacy, Kyungpook National University.
Extraction and isolation – Dried stems of M. alba (30.0 kg) were extracted three times with ethanol (EtOH, 20 L) for 4 hours under reflux conditions. The extract was concentrated under vacuum to yield EtOH soluble residue (1.7 kg). The EtOH soluble residue was dissolved in water and then partitioned sequentially with n-hexane, CH2Cl2, EtOAc, and butanol.
The EtOAc extract (76.8 g) was chromatographed through silica gel vacuum liquid chromatography using a CH2Cl2MeOH elution (gradient 1:0 → 0:1, v/v), resulting in four fractions (1A–1D). Fraction 1B (16.7 g) was then separated using RP-18 CC with MeOH:H2O as an eluent (gradient 4:1 → 0:1, v/v), yielding five fractions (2A–2E). Fraction 2C (1.7 g) was purified by RP-18 CC, eluted with MeOH:H2O (1.5:1, v/v), to isolate compound 1 (51.1 mg). Compounds 2 (20.1 mg) and 3 (117.5 mg) were isolated from fraction 2D (1.3 g) by RP-18 CC, eluting with MeOH:H2O (2.3:1, v/v). Compound 4 (134.0 mg) was isolated from fraction 2E (1.3 g) via RP-18 CC elution with MeOH:H2O (3.2:1, v/v), followed by purification using silica gel CC, eluted with CH2Cl2:MeOH (20:1, v/v). The separated compounds showed high purity, ranging from 92% to 94%.
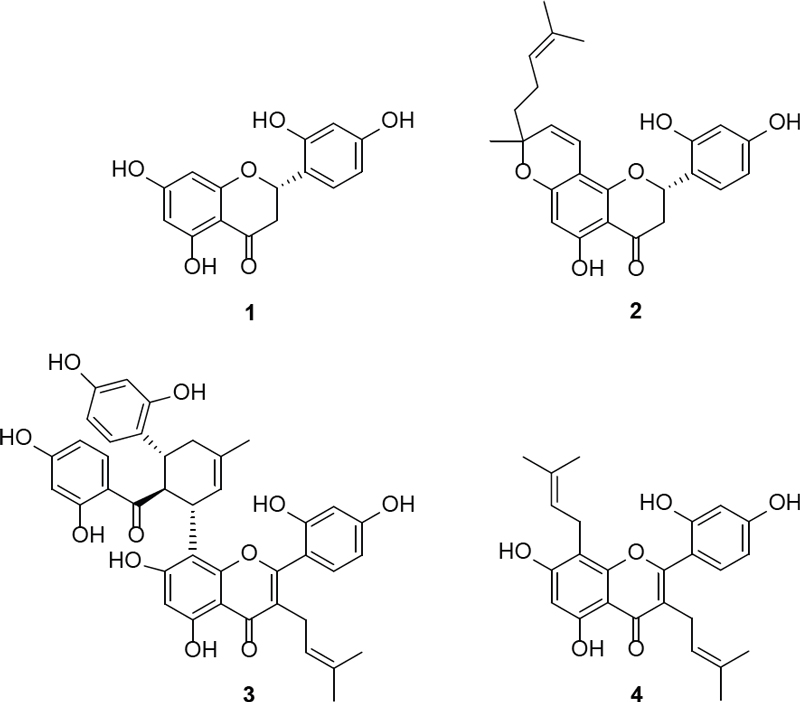
Steppogenin (1) – Amorphous powder; 1H-NMR (CD3OD, 500 MHz): δ 7.22 (1H, d, J=8.0 Hz, H-6′), 6.35 (1H, dd, J=8.0, 2.2 Hz, H-5′), 6.33 (1H, d, J=2.2 Hz, H-3′), 5.91 (1H, d, J=2.2 Hz, H-8), 5.88 (1H, d, J=2.2 Hz, H-6), 5.60 (1H, dd, J=13.1, 2.9 Hz, H-2), 3.06 (1H, dd, J=17.2, 13.1 Hz, H-3a), 2.69 (1H, dd, J=17.2, 2.9 Hz, H-3b); 13C-NMR (CD3OD, 125 MHz): δ 198.5 (C-4), 168.2 (C-7), 165.4 (C-5), 165.3 (C-8a), 159.6 (C-2′), 156.7 (C-4′), 128.9 (C-6′), 117.9 (C-1′), 107.8 (C-5′), 103.4 (C-3′), 103.3 (C-4a), 96.9 (C-6), 96.1 (C-8), 75.9 (C-2), 43.0 (C-3).12
Sanggenol L (2) – Amorphous powder; 1H-NMR (CD3OD, 500 MHz): δ 7.19 (1H, d, J=8.8 Hz, H-6′), 6.56 (1H, d, J=10.4 Hz, H-1″), 6.33 (1H, dd, J=8.8, 2.4 Hz, H-5′), 6.32 (1H, d, J=2.4 Hz, H-3′), 5.86 (1H, br s, H-6), 5.58 (1H, dd, J=13.1, 2.8 Hz, H-2), 5.38 (1H, d, J=10.4 Hz, H-2″), 4.99 (1H, m, H-7″), 2.99 (1H, dd, J=17.2, 13.1 Hz, H-3a), 2.68 (1H, dd, J=17.2, 2.9 Hz, H-3b), 2.00 (2H, m, H-6″), 1.63 (2H, m, H-5″), 1.57 (3H, s, H-10″), 1.49 (3H, s, H-9″), 1.30 (3H, s, H-4″); 13C-NMR (CD3OD, 125 MHz): δ 198.5 (C-4), 164.5 (C-5), 163.4 (C-7), 159.4 (C-4′), 159.1 (C-8a), 156.6 (C-2′), 132.4 (C-8″), 128.8 (C-6′), 125.9 (C-2″), 124.9 (C-7″), 117.6 (C-1′), 116.6 (C-1″), 107.7 (C-5′), 103.5 (C-8), 103.4 (C-4a), 103.3 (C-3′), 96.8 (C-6), 81.7 (C-3″), 75.8 (C-2), 42.9 (C-3), 42.6 (C-5″), 27.5 (C-4″), 25.9 (C-10″), 23.6 (C-6″), 17.8 (C-9″).13,14
Kuwanon G (3) – Amorphous powder; 1H-NMR (CD3OD, 500 MHz): δ 7.26 (1H, d, J=8.2 Hz, H-27), 7.07 (1H, d, J=8.2 Hz, H-6′), 6.67 (1H, d, J=8.2 Hz, H-33), 6.41 (2H, m, H-3′ and H-5′), 6.07 (1H, d, J=2.0 Hz, H-30), 6.01 (1H, dd, J=8.2, 2.4 Hz, H-32), 5.87 (2H, s, H-6 and H-24), 5.83 (1H, d, J=8.2 Hz, H-26), 5.10 (2H, m, H-10 and H-15), 1.55 (3H, s, H-12), 1.40 (3H, s, H-16), 1.37 (3H, s, H-13); 13C-NMR (CD3OD, 125 MHz): δ 210.2 (C-21), 183.8 (C-4), 165.8 (C-23), 165.8 (C-25), 165.6 (C-7), 163.2 (C-4′), 162.4 (C-8a), 161.7 (C-2), 160.9 (C-2′), 157.7 (C-29), 157.7 (C-31), 156.8 (C-5), 134.3 (C-16), 134.1 (C-33), 132.7 (C-11), 132.7 (C-6′), 132.3 (C-27), 124.6 (C-15), 122.9 (C-10), 122.9 (C-28), 121.6 (C-3), 115.9 (C-22), 113.7 (C-1′), 108.5 (C-26), 108.2 (C-8), 107.9 (C-32), 105.6 (C-5′), 103.7 (C-4a), 103.6 (C-24), 103.6 (C-3′), 102.8 (C-30), 98.5 (C-6), 48.5 (C-20), 39.0 (C-18), 39.0 (C-19), 25.9 (C-12), 24.7 (C-9), 23.4 (C-14), 23.1 (C-17), 17.7 (C-13).4,15,16
Kuwanon C (4) – Amorphous powder; 1H-NMR (CD3OD, 500 MHz): δ 7.00 (1H, d, J=8.3 Hz, H-6′), 6.36 (1H, d, J=2.3 Hz, H-3′), 6.33 (1H, dd, J=8.3, 2.3 Hz, H-5′), 6.15 (1H, br s, H-6), 5.07 (1H, m, H-15), 5.02 (1H, m, H-10), 3.23 (1H, d, J=7.0 Hz, H-9a), 3.01 (1H, d, J=7.0 Hz, H-9b), 1.50 (3H, d, J=0.6 Hz, H-18), 1.48 (3H, d, J=0.7 Hz, H-17), 1.46 (3H, s, H-12), 1.30 (3H, s, H-13); 13C-NMR (CD3OD, 125 MHz): δ 183.9 (C-4), 163.5 (C-7), 162.5 (C-4′), 161.6 (C-2), 160.4 (C-8a), 157.7 (C-2′), 156.9 (C-5), 132.6 (C-6′), 132.4 (C-11), 132.0 (C-16), 123.3 (C-15), 122.8 (C-10), 121.3 (C-3), 113.5 (C-1′), 107.8 (C-5′), 107.5 (C-8), 105.3 (C-4a), 103.7 (C-3′), 98.9 (C-6), 25.9 (C-17), 25.8 (C-12), 24.8 (C-9), 22.3 (C-14), 17.7 (C-13), 17.6 (C-18).17
HPLC-DAD instrumentation and conditions – M. alba solutions were analyzed using a Waters Alliance HPLC-DAD system (Waters, MA, USA) equipped with a Waters 1525 Binary HPLC pump, a Waters 2998 Photodiode Array (PDA) detector, and an Agilent Eclipse XD8-C18 column (4.6 × 150 mm, 5 μm). The mobile phase was prepared by mixing 0.1% formic acid in methanol and water, and then filtered through a 0.20 μm PTFE syringe filter before use. Samples were injected at a volume of 10 μL. The UV wavelength settings were 210, 254, and 280 nm. Chromatograms were obtained at a wavelength of 254 nm to enable the simultaneous and thorough visualization of all peaks. The detailed HPLC conditions are presented in (Table 1).
Preparation of standard solutions and analysis samples – For the preparation of calibration curves, four marker compounds (1–4) were each weighed at 1.0 mg and then mixed with HPLC-grade methanol to produce stock solution with a concentration of 1000 μg/mL. These solutions were subsequently diluted to create different concentrations for each marker. M. alba extracts were dissolved in HPLC-grade methanol to achieve a concentration of 20 mg/mL. The resulting solutions were filtered through a 0.20 μm PTFE syringe filter before use.
Method validation – Stock solutions of four marker compounds (1–4) were diluted to concentrations of 5, 10, 50, 100, and 200 μg/mL for HPLC analysis. Calibration curves for the four marker compounds were constructed using the equation: y=ax + b, where a represents the slope of the calibration curve, b is the y-intercept, x is the concentration of the sample, and y is the peak area. Using this data, the correlation coefficient (R2) was calculated, and linearity was determined based on the R2 value of the calibration curve.
The sensitivity of this method was examined by measuring the limit of detection (LOD) and limit of quantitation (LOQ). The LOD represents the lowest concentration that can be reliably determined at a signal-to-noise (S/N) ratio of 3. The determination of the LOQ was calculated using a S/N ratio of 10.18,19
Precision and accuracy were assessed for both intraday (n=5) and inter-day (n=5) measurements by analyzing sets of four marker compounds at different concentrations (50, 150, and 300 μg/mL for each analyte). Precision was evaluated as the relative standard deviation (RSD %), while accuracy was measured as the relative error.
Results and Discussion
The EtOAc extract of M. alba was separated by chromatography techniques, resulting in the isolation of four compounds (Fig. 1). By conducting a comprehensive analysis of NMR spectroscopic data, including 1H and 13C-NMR, and comparing them with literature reports, the chemical structures of isolated compounds were successfully determined as steppogenin (1),12 sanggenol L (2),13,14 kuwanon G (3),4,15,16 and kuwanon C (4).17
The aim of optimizing the HPLC-DAD experiment was to improve the peak resolution of steppogenin and flavonoids from M. alba extracts within a shorter runtime.20 The reversed-phase Agilent Eclipse XD8-C18 column was chosen for the experiment in this study. The mobile phase consisted of a multi-step gradient elution system comprising 0.1% formic acid in methanol (A) and water (B). The separation conditions for the isolated compounds were established at a flow rate of 1.0 mL/min: 0–40 min, 35–100% A; 40–45 min, 100% A; 45–52 min, 100–35% A; and 52–60 min, 35% A. This gradient method facilitated the separation of four distinct peaks within the initial 40 minutes. The Waters 2998 PDA detector was used for detection, with the wavelength set at various settings (210, 254, and 280 nm). Based on the UV spectrum analysis, chromatograms were chosen at 254 nm. Column temperature was observed to have no impact on separation; thus, it was maintained at room temperature (25℃). Confirmation of peak identity of each compound was achieved by comparing retention times and UV spectra with their respective marker solutions. The isolated compounds exhibited retention times of 14.6 min for compound 1, 27.1 min for compound 2, 28.0 min for compound 3, and 30.1 min for compound 4 in the HPLC chromatograms (Fig. 2).
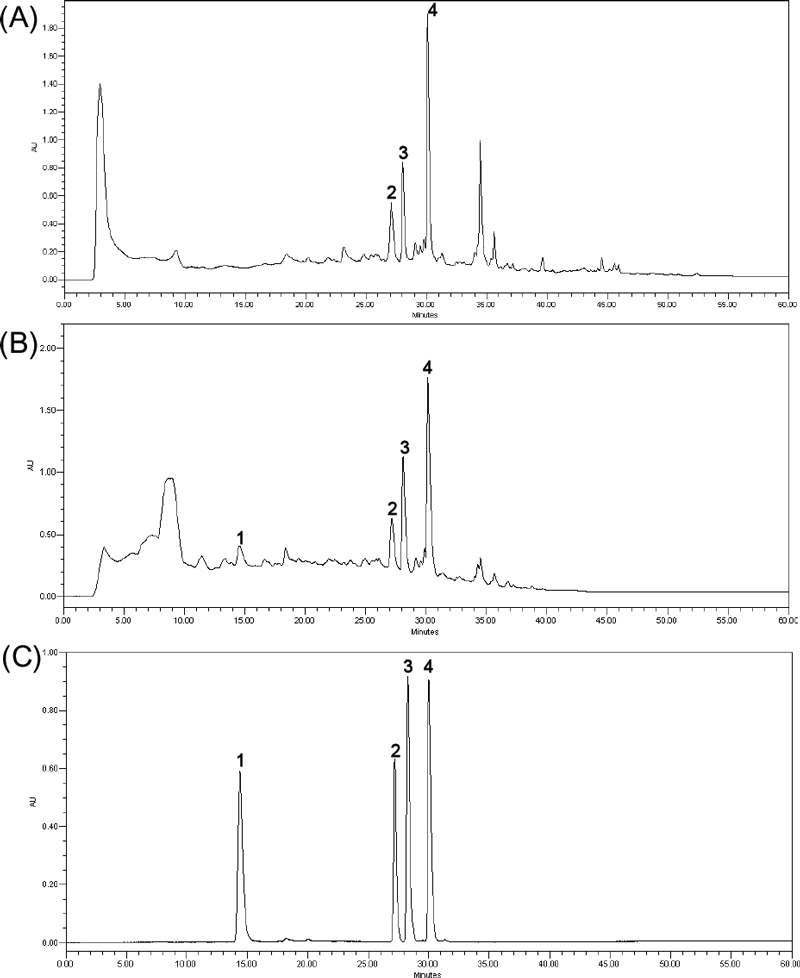
HPLC chromatograms of EtOH (A), EtOAc extracts (B) of M. alba and mixture of four isolated compounds 1–4 (C).
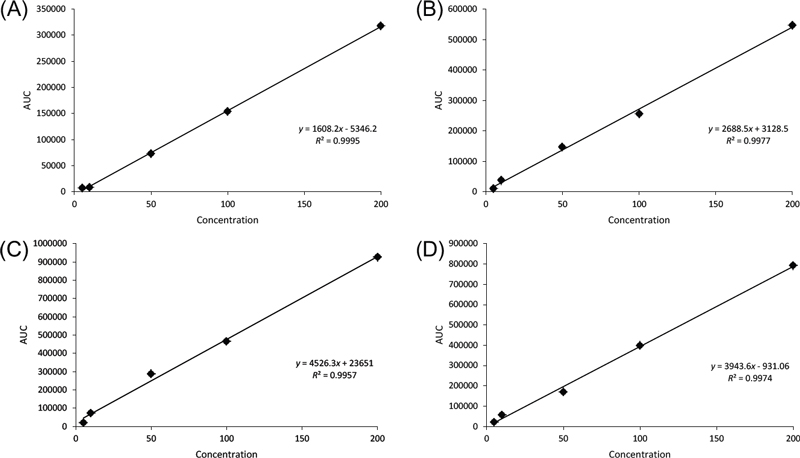
To validate this method, we conducted experiments to assess its linearity, define the limits of detection and quantification, and evaluate precision and accuracy. The linearity of our method was determined by analyzing five concentration levels of each analyte (5, 10, 50, 100, and 200 μg/mL), with each analysis performed in triplicate. The calibration curves for each analyte exhibited excellent linearity across the tested range (R2 > 0.9957) (Fig. 3 and Table 2). The LOD for each marker compound was found to range from 0.006 to 0.018 μg/mL, and the LOQ ranged from 0.020 to 0.061 μg/mL. These findings suggest that our developed method is reliable and demonstrates good sensitivity.

Calibration curves, correlation coefficients (R2), limits of detection (LOD) and limits of quantification (LOQ) of compounds 1–4
To evaluate the accuracy and precision of our developed HPLC-DAD method, both within-day (intra-day) tests (n=5) and intermediate-day (inter-day) tests (n=5) were carried out. The intra-day analysis was conducted five times within a single day, while the inter-day analysis was performed on five different days. As detailed in Table 3, the RSD values obtained for the intra-day tests ranged from 0.22% to 2.27%, and for the inter-day tests, the RSD values ranged from 0.25% to 1.65%. The method demonstrated accuracies within the range of 97.51% to 106.39% for intra-day testing and 97.32% to 105.54% for inter-day testing. Method validation confirmed that the regression equations for the four marker compounds (1–4) were linear, affirming the precision, accuracy, and reliability of this method for quantifying compounds from M. alba.
The contents of all isolated compounds from the stems of M. alba are listed in Table 4. In the EtOH extract, the amounts of compounds 2–4 were found to have average concentrations of 4.01%, 5.33%, and 11.77%, respectively. However, the amount of steppogenin (1) could not be determined. On the other hand, the EtOAc extract was found to have the highest amount of steppogenin at 1.84%. The compounds 2–4 amounts were identified as 2.72%, 6.39%, and 9.08%, respectively.
In this study, steppogenin (1) and three flavonoids (2–4) were isolated from the EtOAc extract of M. alba dried stems. The chemical structures of these isolated compounds were successfully elucidated using spectroscopy techniques, including 1H and 13C-NMR, and subsequently confirmed through comparison with published literature. Additionally, a reliable HPLC-DAD method was developed to simultaneously quantify four marker compounds (1–4). Our developed method significantly enhances the separation efficiency of M. alba constituents, resulting in improved peak resolution. HPLC method validation, including linearity, limits of detection and quantification, precision, and accuracy, highlight the reproducibility and precision of the method. Consequently, a validated analytical method enabled the quantitative analysis of steppogenin and flavonoids from the EtOAc extract of M. alba for the first time, providing valuable foundational data for future investigations on M. alba constituents.
Acknowledgments
This research was supported by the National Research Foundation of Korea (NRF) funded by the Korea government (MSIT) (No. NRF-2020R1A5A2017323). The authors would like to thank Korea Basic Science Institute (KBSI) Daegu Center for the mass spectra measurement service.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
-
Liu, X.-C.; Chen, X.-Q.; Li, R.-T.; Zhang, Z.-J. Biochem. Syst. Ecol. 2023, 107, 104585.
[https://doi.org/10.1016/j.bse.2023.104585]

-
Ha, M. T.; Seong, S. H.; Nguyen, T. D.; Cho, W.-K.; Kim, J. A.; Ma, J. Y.; Woo, M. H.; Choi, J. S.; Min, B. S. Phytochemistry 2018, 155, 114–125.
[https://doi.org/10.1016/j.phytochem.2018.08.001]

-
Dai, S.-J.; Ma, Z.-B.; Wu, Y.; Chen, R.-Y.; Yu, D.-Q. Phytochemistry 2004, 65, 3135–3141.
[https://doi.org/10.1016/j.phytochem.2004.07.010]

-
Chang, Y.-S.; Jin, H.-G.; Lee, H.; Lee, D.-S.; Woo, E.-R. Nat. Prod. Sci. 2019, 25, 268–274.
[https://doi.org/10.20307/nps.2019.25.3.268]

-
Ha, M. T.; Shrestha, S.; Tran, T. H.; Kim, J. A.; Woo, M. H.; Choi, J. S.; Min, B. S. Arch. Pharm. Res. 2020, 43, 961–975.
[https://doi.org/10.1007/s12272-020-01269-4]

-
Zheng, Z.-P.; Tan, H.-Y.; Chen, J.; Wang, M. Fitoterapia 2013, 84, 242–247.
[https://doi.org/10.1016/j.fitote.2012.12.006]

- Chin, H.-S.; NamKung, W. Yakhak Hoeji 2010, 54, 398–402.
-
Cha, S.; Kim, H.-G.; Jang, H.; Lee, J.; Chao, T.; Baek, N.-I.; Song, I.-S.; Lee, Y. M. Phytomedicine 2023, 108, 154513.
[https://doi.org/10.1016/j.phymed.2022.154513]

-
Zhou, Q. Y.-J.; Liao, X.; Kuang, H.-M.; Li, J.-Y.; Zhang, S.-H. Molecules 2022, 27, 5360.
[https://doi.org/10.3390/molecules27175360]

-
Phong, N. V.; Chae, H. Y.; Oanh, V. T.; Min, B. S.; Kwon, M. J.; Kim, J. A. Nat. Prod. Sci. 2023, 29, 171–181.
[https://doi.org/10.20307/nps.2023.29.3.171]

-
Phong, N. V.; Thanh, L. T.; Kwon, M. J.; Lee, Y. M.; Min, B. S.; Kim, J. A. Nat. Prod. Sci. 2023, 29, 349–356.
[https://doi.org/10.20307/nps.2023.29.4.349]

-
Zheng, Z.-P.; Cheng, K.-W.; To, J. T.-K.; Li, H.; Wang, M. Mol. Nutr. Food Res. 2008, 52, 1530–1538.
[https://doi.org/10.1002/mnfr.200700481]

-
Jung, J.-W.; Ko, W.-M.; Park, J.-H.; Seo, K.-H.; Oh, E.-J.; Lee, D.-Y.; Lee, D.-S.; Kim, Y.-C.; Lim, D.-W.; Han, D.; Baek, N.-I. Arch. Pharm. Res. 2015, 38, 2066–2075.
[https://doi.org/10.1007/s12272-015-0613-8]

-
Nam, M. S.; Jung, D.-B.; Seo, K.-H.; Kim, B.-I.; Kim, J.-H.; Kim, J. H.; Kim, B.; Baek, N.-I.; Kim, S.-H. Phytother. Res. 2016, 30, 90–96.
[https://doi.org/10.1002/ptr.5505]

-
Oshima, Y.; Konno, C.; Hikino, H.; Matsushita, K. Tetrahedron Lett. 1980, 21, 3381–3384.
[https://doi.org/10.1016/S0040-4039(00)78694-1]

-
Hsu, J.-H.; Yang, C.-S.; Chen, J.-J. Antioxidants 2022, 11, 2222.
[https://doi.org/10.3390/antiox11112222]

-
Guo, S.; Liu, L.; Zhang, S.; Yang, C.; Yue, W.; Zhao, H.; Ho, C.-T.; Du, J.; Zhang, H.; Bai, N. Food Funct. 2019, 10, 6915–6926.
[https://doi.org/10.1039/C9FO01457H]

-
Kim, E.-N.; Lee, D.-E.; Lee, W. J.; Kim, T. Y.; Roh, Y.-S.; Jeong, G.-S. Kor. J. Pharmacogn. 2023, 54, 191–196.
[https://doi.org/10.22889/KJP.2023.54.4.191]

- Nguyen, D. H.; Le, D. D.; Ma, E. S.; Min, B. S.; Woo, M. H. Nat. Prod. Sci. 2020, 26, 326–333.
-
Yang, W.; Ma, C. J. Nat. Prod. Sci. 2023, 29, 305–311.
[https://doi.org/10.20307/nps.2023.29.4.305]