
Characterization of Glycosyl Inositol Phosphoceramides from Panax ginseng using Nanospray Tandem Mass Spectrometry
Abstract
Korean ginseng (Panax ginseng C. A. Meyer) is one of the most popular medicinal herbs in the world. This plant is known to have many health benefits and contain a wide variety of bioactive components. However, the knowledge on its lipid compound is still far from being fully explored. Although glycosyl inositol phosphoceramides (GIPCs) are the main sphingolipids in plant tissues, GIPCs of P. ginseng are unknown. The present study employed nanoESI-MSn, which generated characteristic fragmentation pattern that were used for the structural identification of P. ginseng GIPCs. In addition to detecting a typical mass fragmentation pattern for GIPC in positive ion mode, novel fragmentation correlating with cleavage of the last carbohydrate and fatty acyl chain of the ceramide moiety was identified. In total, 42 GIPC species were detected in P. ginseng. The major P. ginseng GIPC structure was hexose (R1)-hexuronic acid-inositol phosphoceramide, with three types of R1 (amine, N-acetylamine, or hydroxyl). The most intense peak was found at m/z 1136.3 ([M+H]+ ion), corresponding to a GIPC (d18:0/h16:0, R1 = OH). Only three GIPC subtypes showed significantly different levels in five- and six-year-old P. ginseng tap roots.
Keywords:
Panax ginseng, Glycosyl inositol phosphoceramides, NanoESI-MSIntroduction
Sphingolipids are recognized as ubiquitous constituents of plasma cell membrane, the tonoplast, and other plant cell endomembranes with diverse biological functions.1–3 In particular, two complex sphingolipids have been associated with plant membrane lipid rafts, which are essential for the trafficking and sorting of vital proteins: these are glycosyl inositol phosphoceramides (GIPCs) and glucosylceramides (glucocerebroside).4 GIPCs are only encountered in plants and fungi, and not in animals or bacteria. These lipids are the predominant sphingolipids in plant membranes.5 GIPCs are believed to be involved in diverse cellular functions. Wang et al. demonstrated that inositol phosphoryl ceramide (IPC), the core compartment of GIPC, regulate programmed cell death associated with resistance to pathogens.6
Only a few GIPCs have been fully characterized to date, despite this being one of the most abundant and earliest identified classes of plant sphingolipids.7,8 This is mainly due to their large hydrophilic polar head, which results in relatively poor extraction yield using a classical phase partition technique with a chloroform/methanol/water mixture.9,10 Markham et al. reported that extraction with 2-propanol/hexane/water (55:20:25, v/v/v) at 60°C provided a useful and efficient method for extracting sphingolipids including GIPCs, from A. thaliana leaves.11 Buré et al. have optimized the original protocol for GIPC extraction reported by Carter and Koob, where the removal of protein and use of water/butan-1-ol (1:1, v/v) produced optimal GIPC recovery.8,9
Used in combination with selective extraction, mass spectrometric techniques play a crucial role in the analysis and structural characterization of these complex sphingolipids. The GIPCs in A. thaliana leaves, tomato, and soybeans were determined by high-performance liquid chromatography coupled to tandem mass spectrometry.10,11 In addition, GIPCs in A. thaliana leaves/cell and tobacco BY-2 cell were identified by matrix-assisted laser desorption/ionization (MALDI) MS/MS.12 Cacas et al. elucidated the structure of GIPCs from 23 plant species representing various phylogenetic groups using a MALDI-MS/MS platform.13 In addition, GIPC profiling was performed for spinach, white cabbage, sunflower seeds, and soybeans using a linear ion trap-orbitrap (LTQ Orbitrap) mass spectrometer.14
The core structure of GIPC is composed of a ceramide backbone linked to a glucuronic acid-inositol phosphate group. Additional diverse saccharide molecules can be attached to this core structure.12 The ceramide backbone, consists of a fatty acid (FA) bound to a long-chain base (LCB) via an amide linkage.1–3 Plant sphingolipid FAs are often α-d-hydroxylated in position 2 and generally range in chain length from 16 to 26 carbon atoms, which can be either saturated or monounsaturated, with a cis-ω9 double bond. Plant LCBs contain 18 carbon atoms and are characterized by the presence of either two (represented as the prefix of ‘d’) or three (represented as the prefix of ‘t’) hydroxyl groups. The LCBs found in plant sphingolipids are sphinganine (or dihydrosphingosine; d18:0), sphingenine (d18:1), sphigadiene (d18:2), 4-hydroxysphinganine (or phytosphingosine; t18:0), and 4-hydroxysphingenine (t18:1). The predominant LCB structures of GIPCs are t18:0 and t18:1.12 The proportion of these LCBs in the ceramide backbone can vary widely between different organs in a single species, and between different species.1
There are several lines of evidence indicating that dietary sphingolipids produce beneficial effects on human health. Berra et al. reported that sphingomyelin (a ceramide molecule attached to a phosphocholine head group) supplementation reduced colonic adenocarcinomas in CF1 mice.15 In addition, glycosphingolipids can protect against bacteria toxins and infection because many bacteria and virus utilize glycosphingolipids to adhere to cells; dietary sphingolipids compete for these cellular binding sites and thus facilitate the removal of pathogenic organisms from the intestine.16
In order to understand the role of GIPCs in plant system, as well as their pharmacological effects in humans, it is important to characterize diverse sphingolipids qualitatively and quantitatively. Although research has been conducted to examine GIPCs in model plants such as A. thaliana and tobacco, there have been few studies of GIPCs in other herbal plants.
The objective of this work was the design of a nanoelectrospray tandem mass spectrometry (nanoESI-MSn) approach that could provide a quick and complete description of GIPCs in the P. ginseng tap root. In addition, the GIPC contents were compared in five-and six-year-old P. ginseng roots in order to investigate age-related changes in these molecules.
Experimental
Sample preparation – Ginseng roots (Panax ginseng C.A. Meyer cv. Chunpoong) were prepared as previously described.17 The tap roots from five- and six-year-old P. ginseng were used for GIPC extraction according to a method adapted from Markham and Jaworski.11 Briefly, freeze-dried powder was sieved using a 200 mesh sieve. An accurately weighed 30 mg sample of dry powder was weighed into a 10 mL glass centrifuge tube, to which 3 mL extraction solvent [isopropanol/hexane/water (55:20:25, v/v/v)] was added. The tubes were capped and vortexed for 10 s, sonicated for 10 min, and then incubated at 60°C for 15 min. After centrifugation at 500 g for 10 min, the supernatant was decanted to a second tube and the pellet was extracted once more with 3 mL extraction solvent. After a second incubation at 60°C for 15 min and centrifugation as before, the supernatants were combined and dried under a stream of nitrogen. The dried extract was dissolved in 1 mL reconstitution solvent [acetonitrile/water (95:5, v/v)]. This was then spun at 500 g for 10 min to remove any insoluble material and passed through a 0.2 μm PTFE syringe filter (Whatman, Maidstone, UK), 0.4 mL of the filtrate was then evaporated under nitrogen gas. Dried lipid extracts were dissolved in 0.4 mL of acetonitrile/water (95:5, v/v) containing 10 mM ammonium acetate buffer solution, and 40 μL of this solution was loaded into a 96 well plate (Eppendorf, Hamburg, Germany) and sealed with aluminum foil (Eppendorf).
Analysis of GIPCs – Lipids were analyzed in positive ion mode on a linear ion-trap mass spectrometer (LTQ-XL, ThermoFisher Scientific, San Jose, CA, USA) equipped with an automated nanoinfusion/nanospray source (TriVersa NanoMate System, Advion Biosciences, Ithaca, NY, USA). The ionization voltage and gas pressure were set to 1.4 kV and 0.4 psi, respectively. The ion source was controlled using Chipsoft software (version 8.3.1, Advion Biosciences). The 96 well plate was placed on a NanoMate cooling plate set to 4°C. Ten microliters of each sample was aspirated and infused into a mass spectrometer via an Advion ESI chip with 5.5 μm (inner diameter) emitter nozzles. The samples were analyzed in triplicate by nanoESI-MSn. To eliminate potential bias associated with analysis order, the analysis sequence of the samples was randomized.
Full-scan spectra were collected in mass-to-charge ratio (m/z) ranges of 1100–1400 using the positive ion mode. The mass spectrum of each sample was acquired in profile mode over a 2 min period. The capillary temperature was set to 200°C and the capillary and tube-lens voltages were set to 31 V and 250 V, respectively, in positive-ion mode. The target automatic-gain-control values for full MS and multistage MS were 30,000 and 1,000, respectively. To identify GIPCs, candidate molecule ions ([M+H]+) in the lipid extract were subjected to product ion MS/MS scanning. The normalized collision energy was set to 50%, with an isolated width of 1.5 m/z units and a charge state of 1.
Identification and data processing – Lipids were tentatively identified based on their specific MS/MS fragmentation patterns, which have previously been reported in the literature, and an in-house MS/MS library.9–11 The proposed fragmentation patterns were generated using MassFrontier software (version 7.0, HighChem, Bratislava, Slovakia). The raw data files acquired from analyzed samples were processed with Expressionist MSX software (version 2013.0.39, Genedata, Basel, Switzerland). The spectral scans were averaged prior to spectrum smoothing, m/z alignment, and baseline subtraction. The significance of any differences in GIPC levels was tested by student’s t-test (at a threshold of p < 0.05) using SPSS Statistics software (version 21, IBM, Somers, NY, USA).
Results and Discussion
The targeted MS/MS analysis by nanoESI-MSn provided specific fragment ion information for GIPCs, which contain a polar head group and multiple LCBs and FAs with varying degrees of saturation and hydroxylation. GIPC standards are not commercially available and the fragmentation patterns were therefore compared to those presented by previous studies.9–11 All GIPC species in this study were detected as mainly protonated form [M+H]+, although the presence of sodium, ammonium, potassium, and lithium adducts was also reported in other studies.10,11,18 Product ion scan spectra of these protonated ions [M+H]+ provided informative fragmentation patterns for the structural identification of GIPC species. Using the information provided from these scans, the molecular structure of the major charged sphingolipid from P. ginseng was proposed to be hexose(R1)-hexuronic acid-inositol-phosphoceramide, with R1 being an amine, an N-acetylamine group or a hydroxyl (Fig. 1). The exact identity of the hexose and hexuronic acid could not be assigned based on the mass spectrum.10
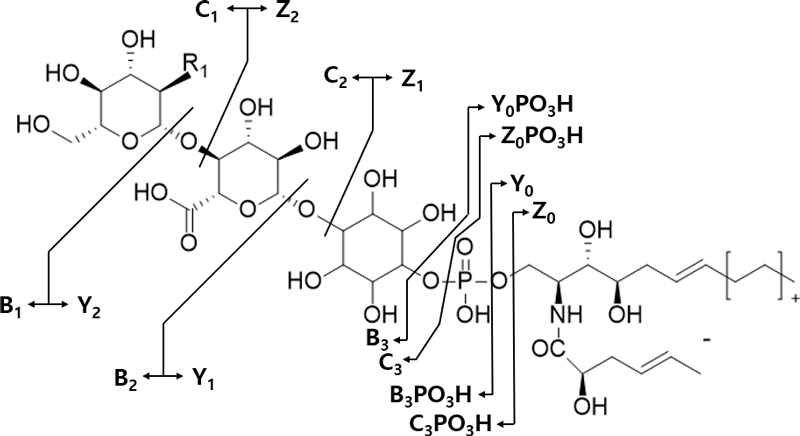
Fragmentation pattern of GIPC precursor ions in the positive ion mode, adapted from Markham et al. (2006)
Fragment ions were designated according to the nomenclature proposed by Costello and Vath (1990) and Levery et al. (2001); characteristic ions are presented in a fragmentation scheme in Fig. 1.18,19 As an example, the fragment ion spectra for m/z 1119.9 [with a d18:0/c16:0 ceramide moiety and R1 as an amine; GIPC (d18:0/c16:0, R1 = NH2)], m/z 1306.0 GIPC (t18:0/h24:0, R1 = NHCOCH3), and m/z 1206.6 GIPC (t18:0/h20:1, R1 = OH) are presented in Fig. 2. The presence of an hexuronic acid-inositol-phosphate moiety was proved by the detection of C3PO3-B1 ions (m/z 437.0). The R1 of hexose was evidenced by the detection of the Y2 ion, which has the mass difference from precursor ion [M+H]+ by 161, 203, 162 Da, corresponding to glucosamine, N-acetylglucosamine and glucuronic acid, respectively. The ceramide moiety was identified by the Y0PO3 or Z0PO3 ions.
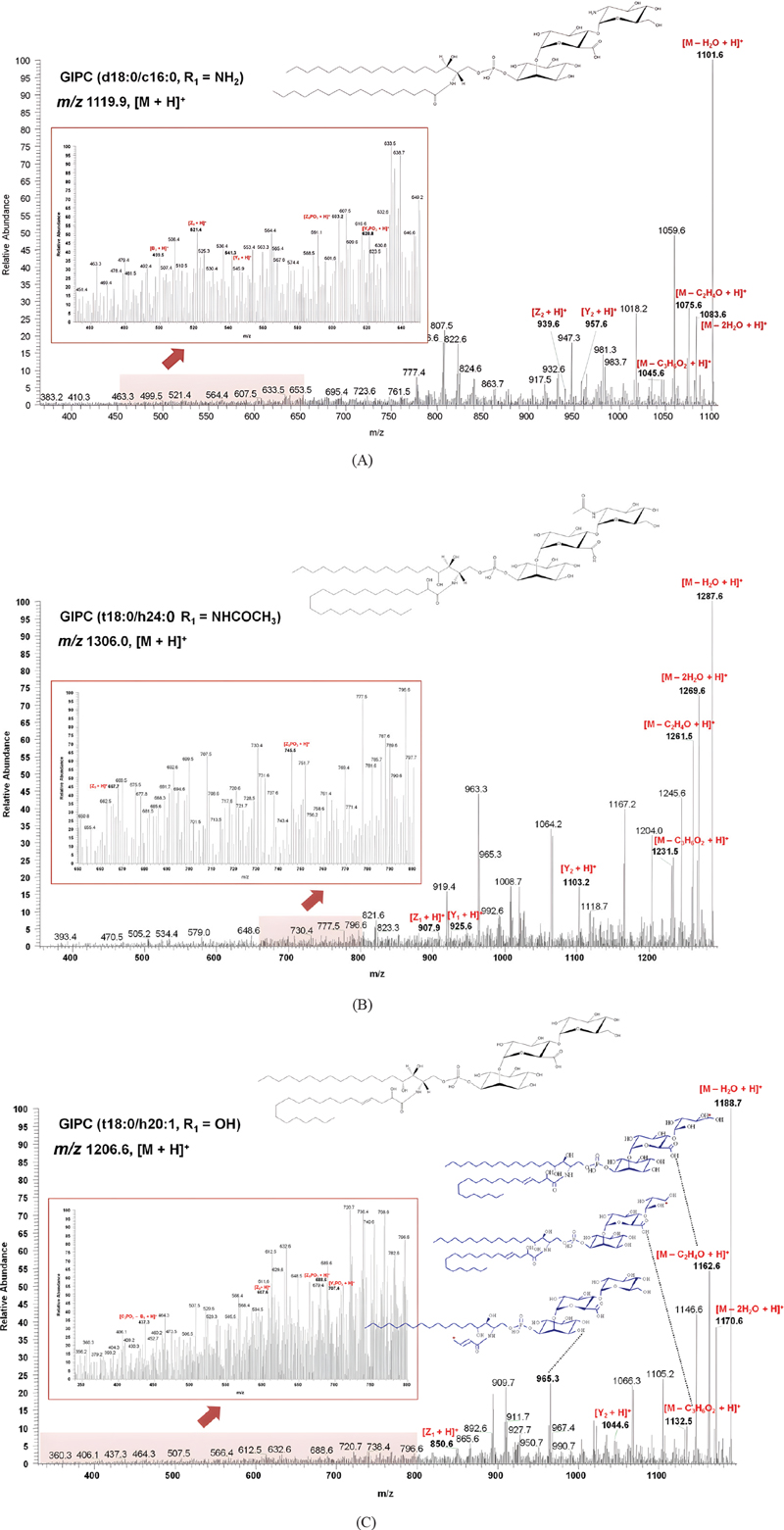
The representative MS/MS spectra of (A) m/z 1119.9, glycosyl inositol phosphoryl ceramide (GIPC) (d18:0/c16:0, R1 = NH2), (B) m/z 1306.0, GIPC (t18:0/h24:1, R1 = NHCOCH3), and (C) m/z 1206.6, GIPC (t18:0/h20:1, R1 = OH) from P. ginseng. The nomenclature for GIPC fragmentation is illustrated in Fig 1. The prefixes “d” and “t” designate a di- and tri-hydroxy sphingoid backbone, respectively. The prefixes “h” and “c” designate hydroxylated and non-hydroxylated fatty acids, respectively.
The MS/MS spectra included a series of fragment ion masses with differences of 18, 36, 44, and 74 Da, which originated from the loss of the one- and two-water molecules, C2H4O and C3H6O2, respectively, from their protonated molecular ion. The losses of C2H4O and C3H6O2 likely reflected the cleavage of the last hexose of GIPC. The putative fragmentation mechanism was illustrated in Fig. 3A and B. A previous review article described the loss of C3H6O2 due to cleavage of the C-3 and C-5 positions in the last sugar unit; however, this fragment ion has not been previously demonstrated using MS/MS.12 In addition, this is the first description, to our knowledge, of the loss of C2H4O due to cleavage at the C-4 and C-5 positions.
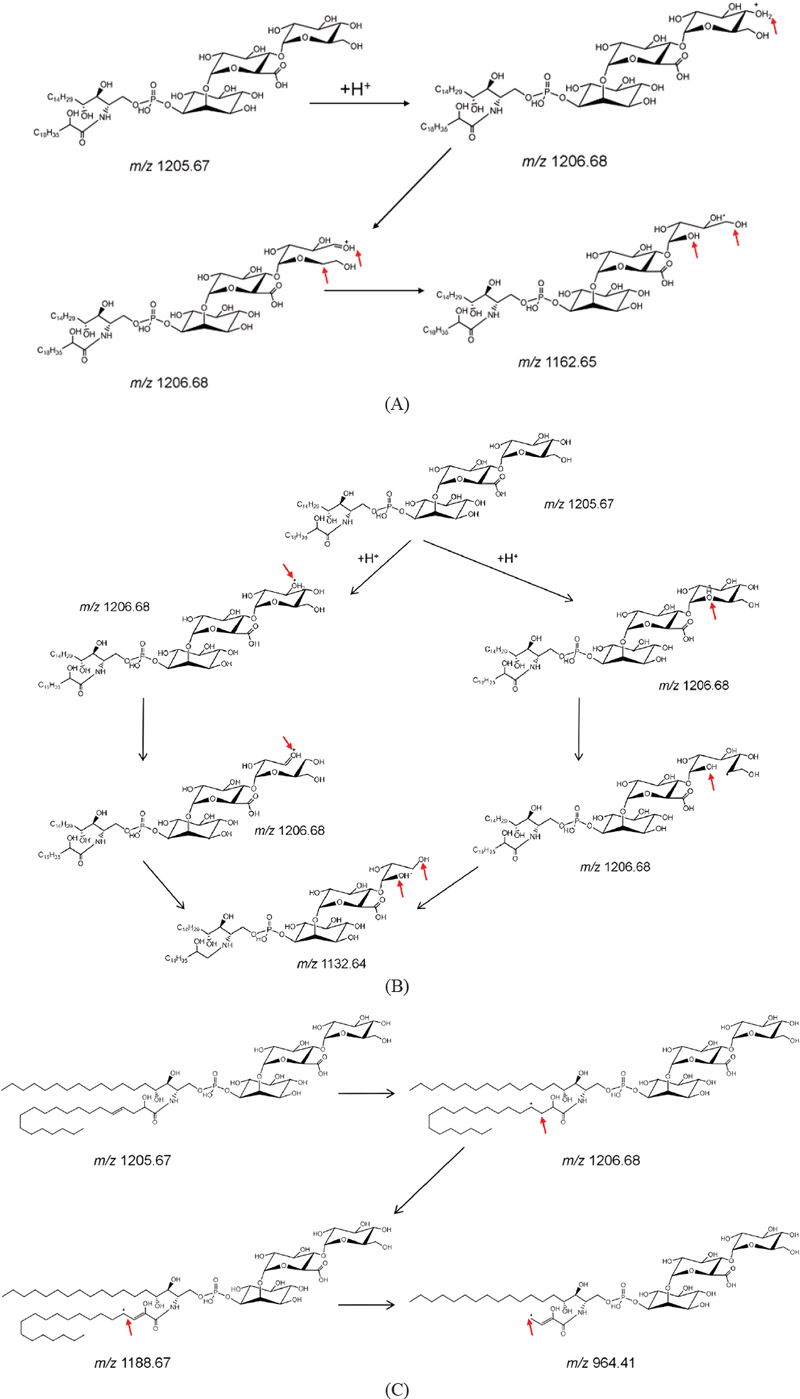
Proposed fragmentation pathways of the fragment at (A) m/z 1162.6, (B) m/z 1132.5, and (C) m/z 965.3 from glycosyl inositol phosphoryl ceramide (GIPC) (t18:0/h20:1, R1 = OH) following collision-induced dissociation in positive ion mode. This scheme was predicted by MassFrontier software.
The characteristic LCB ions were found in all GIPCs and interestingly, these m/z values were not affected by the range of FA chains. These ions have not previously been reported by positive ion fragmentation studies. Table 1 lists the ions observed in GIPCs of P. ginseng and the proposed fragmentation mechanism was presented in Fig. 3C. These ions were used to select the main precursor ion from isobaric species. As an example, the fragment ion spectrum for m/z 1259.9 [GIPC (t18:1/h24:1, R1 = NH2)] is presented in Fig. 4. This species included isobaric compounds: GIPC (d18:0/c26:0, R1 = NH2), GIPC (d18:0/h22:1, R1 = NHCOCH3), and GIPC (t18:1/c22:0, R1 = NHCOCH3). For the m/z 961.7 ion, the characteristic ion from GIPC (t18:1/h24:1, R1 = NH2) showed the highest sensitivity of these characteristic ions from other isobaric compounds, and the m/z 1259.9 precursor ions was therefore determined as GIPC (t18:1/h24:1, R1 = NH2).

Characteristic fragment ions according to long-chain bases (LCBs) of GIPC identified from P. ginseng. The columns represent LCB types and the rows represent fatty acids (FAs) types.
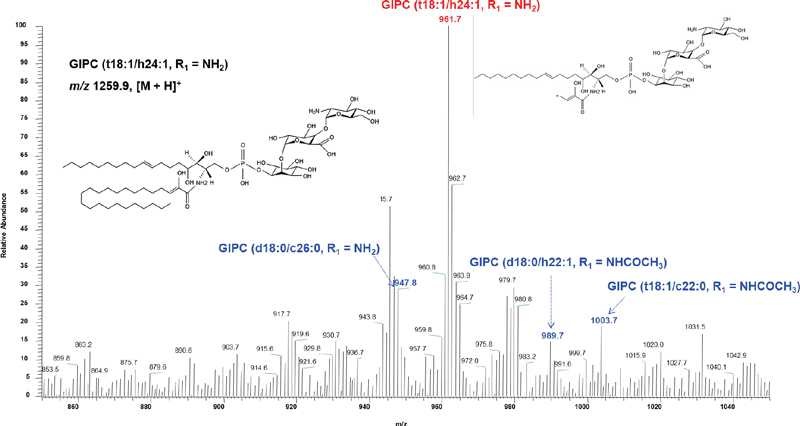
The MS/MS spectrum of m/z 1259.9 with m/z ranges of 1100–1400 in positive ion mode. GIPC, glycosyl inositol phosphoryl ceramide. The prefixes “d” and “t” designate a di- and tri-hydroxy sphingoid backbone, respectively. The prefixes “h” and “c” designate hydroxylated and non-hydroxylated fatty acids, respectively.
The direct infusion of lipid extracts into a tandem mass spectrometer has been used for GIPC analysis in various plants.9–11,13,14 However, we are not aware of any previous studies using the nanoESI-MSn method to profile GIPCs in P. ginseng. These nanoESI-MS spectra are shown and the assigned structures are listed in Table 2. In plants, a number of saccharides are typically linke to inositol; for instance, up to 14 saccharides were found in tobacco leaves.20 In the present study, P. ginseng GIPCs were modeled on the structure of A. thaliana GIPCs, which contain two glycosylated residues linked to inositol, described as hexose(R1)-hexuronic acid-inositol-phosphoceramide.
Based on established GIPC mass fragmentation patterns, a total of 42 GIPC species were identified. In P. ginseng; these had three types of hexose R1 group (amine, acetylamine or hydroxyl). Previous studies have shown that A. thaliana leaves and cells contain hexose with an hydroxyl R1 group [hexose(OH)], soybean and tomato plants contain hexose(NHCOCH3), while Nicotiana tabacum cells contain both hexose (NH2) and hexose(NHCOCH3).10,11 Therefore, the point that P. ginseng GIPCs contain all these three types of R1 is the signature of P. ginseng GIPCs.
Since pure reference compounds were not available at this stage of the work, precise quantification of GIPC could not be conducted by full scan nanoESI-MS. However, Fig. 5. provides an overview of the GIPCs present in five-and six-year-old P. ginseng tap roots, with their various combinations of LCBs and FAs, and a comparison of the GIPC intensities. Only three GIPC species [GIPC (d18:2/h26:0, R1 = NHCOCH3), GIPC (d18:2/h20:1, R1 = OH), and GIPC (t18:0/c22:0, R1 = OH)] showed significantly different levels in five- and six-year-old P. ginseng tap roots (p < 0.05). This result contrasted with those of a previous study of glycerolipids, which showed remarkable differences in five- and six-year-old P. ginseng.17
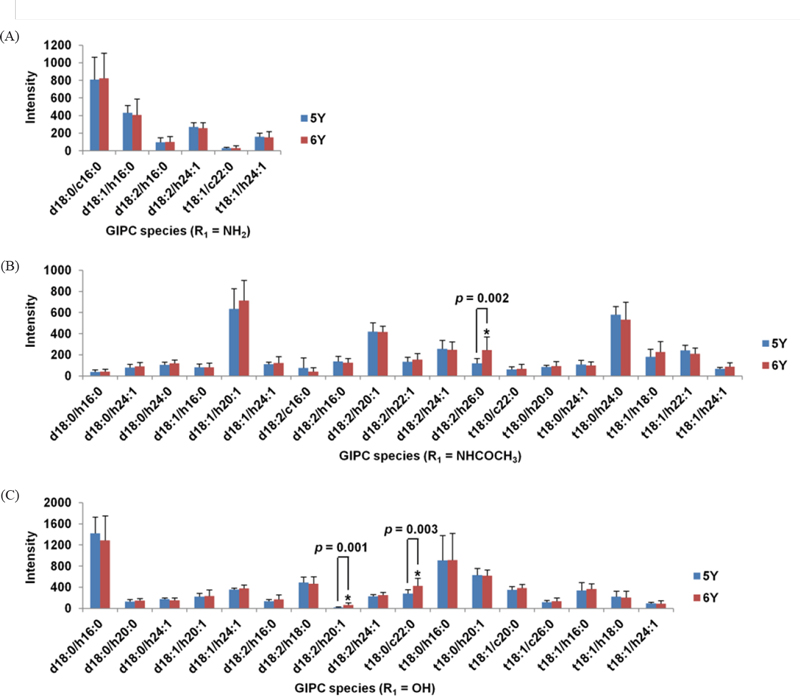
Nanoelectrospray tandem mass spectrometry intensity of detected subspecies of glycosyl inositol phosphoryl ceramides (GIPCs) from five-year-old (5Y) and six-year-old (6Y) P. ginseng tap roots, grouped according to hexose(R1) type: (A) amine, (B) N-acetylamine, and (C) hydroxyl group. The prefixes “d” and “t” designate a di- and tri-hydroxy sphingoid backbone, respectively. The prefixes “h” and “c” designate hydroxylated and non-hydroxylated fatty acids, respectively. Significant differences between five- and six-year-old P. ginseng tap roots are indicated, Student’s t test.
The major FAs of GIPCs in P. ginseng appeared to be h16:0 and h24:1. Hydroxylated FAs are more prevalent than non-hydroxylated FA (Sperling and Heinz 2003).3 In particular, saturated C16, C20, C22, C24 α-hydroxylated FAs were previously reported to be the most abundant, while monounsaturated very long-chain fatty acids ranging from C22 to C26 occurred less frequently.21 In this study, the h26:0 was exclusively observed in acetylamine group.
In LCB profiles, GIPC species containing d18:2 and t18:1 were frequent in the present study. Previous studies have shown that the proportion of d18:2 varied between taxonomic lines. For instance, the plants belong to the family Solanaceae (tomato and tobacco) have a large amount of d18:2, the family Fabaceae (pea and soybean) have a medium amount of d18:2, and the family Brassicaceae (including Arabidopsis) contain very low to negligible levels of d18:2. This could reflect the different activity of Δ4 desaturase in these species.10 Therefore, an analysis of GIPC in A. thaliana did not identify d18:2 as an LCB form within their GIPC molecules. However, this GIPC form was the most prevalent in the present study of P. ginseng (12 molecules, see Table 9).
The most intense peaks were found at m/z 1136.3 ([M+H]+ ion), corresponding to a GIPC (d18:0/h16:0, R1 = OH). Less intense species at m/z 1152.3 corresponded to GIPC (t18:0/h16:0, R1 = OH) and one at m/z 1119.9 corresponded to GIPC (d18:0/c16:0, R1 = NH2) (Fig. 14). This differed from A. thaliana, where GIPC (t18:1/h24:1, R1 = OH) was the most intense peak.9,10
We successfully developed a nanoESI-MSn method for structural elucidation of GIPC species in P. ginseng tap roots and used this to identify a total of 42 GIPCs in these samples. While in other plants, the R1 of the last hexose structure is a hydroxyl, an amine or a N-acetylamine, P. ginseng GIPCs included all three types of R1. In addition to the typical GIPC fragmentation pattern, two types of fragments observed suggested cleavage of the last sugar ring. Although the presence of isobaric species can represent a complicating factor for GIPC identification following direct MS analysis without prior chromatographic separation, isobaric GIPC species were differentiated in the present study using the fragment ion information that resulted from cleavage of the fatty acyl chain.
The multiple health-promoting properties of P. ginseng are believed to result from synergistic effects of various plant constituent, but there is a paucity of studies on P. ginseng lipids, including GIPCs. As the exact biological roles of GIPCs in plants and humans remain unknown, the GIPC profile could provide fundamental information for elucidating the mechanisms underlying the bioactivities of P. ginseng. In addition, this platform could provide a useful method for characterizing GIPC species within other herbal resources.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP) [NRF-2022R1A2C1005245].
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
-
Chen, M.; Cahoon, E.-B.; -García, M.-S.; Plasencia, J.; Ruíz, M.-G. In Lipids in Photosynthesis. Advances in Photosynthesis and Respiration. Wada, H., Murata, N. Ed; Springer; Amsterdam, 2009, pp 77–115.
[https://doi.org/10.1007/978-90-481-2863-1_5]

-
Lynch, D.-V.; Dunn. T.-M. New. Phytol. 2004, 161, 677–702.
[https://doi.org/10.1111/j.1469-8137.2004.00992.x]

- Sperling, P.; Heinz, E. Biochim. Biophys. Acta. 2003, 71, 1–15.
-
Borner, G. H. H.; Sherrier, D. J.; Weimar, T.; Michaelson, L. V.; Hawkins, N. D.; Macaskill, A.; Napier, J. A.; Beale, M. H.; Lilley, K. S.; Dupree, P. Plant. Physiol. 2005, 137, 104–116.
[https://doi.org/10.1104/pp.104.053041]

-
Cacas, J.-L.; Furt, F.; Le Guédard, M.; Schmitter, J.-M.; Buré, C.; Gerbeau-Pissot, P.; Moreau, P.; Bessoule, J.-J.; Simon-Plas, F.; Mongrand, S. Prog. Lipid Res. 2012, 231, 272–299.
[https://doi.org/10.1016/j.plipres.2012.04.001]

-
Wang, W.; Yang, X.; Tangchaiburana, S.; Ndeh, R.; Markham, J. E.; Tsegaye, Y.; Dunn, T. M.; Wang, G.-L.; Bellizzi, M.; Parsons, J. F.; Morrissey, D.; Bravo, J. E.; Lynch, D. V.; Xiao, S. Plant Cell 2008, 20, 3163–3179.
[https://doi.org/10.1105/tpc.108.060053]

-
Carter, H. E.; Gigg, R. H.; Law, J. H.; Nakayama, T.; Weber, E. J. Biol. Chem. 1958, 50, 1309–1314.
[https://doi.org/10.1016/S0021-9258(18)49332-5]

-
Carter, H. E.; Koob, J. L. J. Lipid Res. 1969, 52, 363–369.
[https://doi.org/10.1093/ajcp/52.3_ts.363]

-
Buré, C.; Cacas, J.-L; Wang, F.; Gaudin, K.; Domergue, F.; Mongrand, S.; Schmitter, J.-M. Rapid Commun. Mass. Spectrom. 2011, 25, 3131–3145.
[https://doi.org/10.1002/rcm.5206]

-
Markham, J. E.; Li, J.; Cahoon, E.-B.; Jaworski, J. G. J. Biol. Chem. 2006, 281, 22684–22694.
[https://doi.org/10.1074/jbc.M604050200]

-
Markham, J. E.; Jaworski, J. G. Rapid Commun. Mass Spectrom. 2007, 21, 1304–1314.
[https://doi.org/10.1002/rcm.2962]

-
Buré, C.; Cacas, J.-L.; Mongrand, S.; Schmitter, J.-M. Anal. Bioanal. Chem. 2014, 406, 995–1010.
[https://doi.org/10.1007/s00216-013-7130-8]

-
Cacas, J.-L.; Buré, C.; Furt, F.; Maalouf, J.-P.; Badoc, A.; Cluzet, S.; Schmitter, J.-M.; Antajan, E.; Mongrand, S. Phytochemistry 2013, 96, 191–200.
[https://doi.org/10.1016/j.phytochem.2013.08.002]

-
Blaas, N.; Humpf, H.-U. J. Agric. Food Chem. 2013, 61, 4257–4269.
[https://doi.org/10.1021/jf4001499]

-
Berra, B.; Colombo, I.; Sottocornola, E.; Giacosa, A. Eur. J. Cancer Prev. 2002, 11, 193–197.
[https://doi.org/10.1097/00008469-200204000-00013]

-
Fantini, J.; Hammache, D.; Delézay, O.; Yahi, N.; André-Barrès, C.; Rico-Lattes, I.; Lattes, A. J. Biol. Chem. 1997, 272, 7245–7252.
[https://doi.org/10.1074/jbc.272.11.7245]

-
Kim, S.-H.; Shin, Y.-S.; Choi, H.-K. Anal. Bioanal. Chem. 2016, 408, 2109–2121.
[https://doi.org/10.1007/s00216-016-9314-5]

-
Levery, S. B.; Toledo, M. S.; Straus, A. H.; Takahashi, H. K. Rapid Commun. Mass Spectrom. 2001, 15, 2240–2258.
[https://doi.org/10.1002/rcm.505]

-
Costello, C. E.; Vath, J. E. Meth. Enzymol. 1990, 193, 738–768.
[https://doi.org/10.1016/0076-6879(90)93448-T]

-
Kaul, K.; Lester, R. L. Plant Physiol. 1975, 55, 120–129.
[https://doi.org/10.1104/pp.55.1.120]

-
Imai, H.; Yamamoto, K.; Shibahara, A.; Miyatani, S.; Nakayama, T. Lipids 2000, 35, 233–236.
[https://doi.org/10.1007/BF02664774]
