PTP1B Inhibitory Activity of Flavonoids from the Roots of Astragalus membranaceus Bunge
Abstract
The roots of Astragalus membranaceus Bunge have long been used in herbal medicine for their diverse biological activities. Notably, its potential anti-diabetic properties have been extensively studied, highlighting promising therapeutic prospects. In this study, we conducted a comprehensive investigation focusing on flavonoid components from the roots of A. membranaceus and their PTP1B inhibitory activity. As a result, we isolated a total of 24 flavonoids, among which formonentin (1), pratensein (3), and vesticarpan (19) emerged as the most potent inhibitors against PTP1B with IC50 value of 10.9 ± 1.09 μM, 10.0 ± 1.71 μM, and 10.3 ± 1.31 μM, respectively. Additionally, through the enzyme kinetic analysis, the inhibition mode of compound 19 was determined as a competitive inhibitor, with Ki value of 7.6 ± 1.17 μM. Furthermore, the molecular docking simulation elucidated the binding mechanism of compound 19 with PTP1B, mainly through van der Waals forces and hydrogen bonds. This study highlights the PTP1B inhibitory potential of the flavonoid constituents derived from the roots of A. membranaceus. Moreover, discovering vesticarpan (19) as a novel PTP1B inhibitor provides a significant foundation for further investigations to develop innovative therapeutic strategies for diabetes treatment.
Keywords:
Astragalus membranaceus, Fabaceae, Flavonoid, PTP1B, DiabetesIntroduction
Diabetes mellitus (DM) is a chronic, non-contagious condition with elevated morbidity levels and costly treatment demands.1 It is classified into two primary forms: type 1 diabetes (T1D), characterized by deficient insulin production, and type 2 diabetes (T2D), where insulinresponsive cells exhibit resistance. Both types of diabetes demand prolonged treatment, often leading to unforeseen complications and adverse effects on the body.2 Furthermore, diabetes is recognized as a prevalent risk factor for other chronic conditions, including vision impairments, cardiovascular issues, and neuropathy.3–5 Therefore, diabetes is considered as a high-risk disease, emphasizing the importance of ongoing research to develop novel therapies. Insulin injection remains the principal treatment for T1D, while T2D is treated using various therapeutic methods targeting specific proteins in the insulin pathway.6
Protein-tyrosine phosphatase 1B (PTP1B) has gained attention as a potential therapeutic target for T2D treatment. To date, it has been identified as a therapeutic target for a range of diseases, including diabetes, obesity, cardiovascular disorders, and hematopoietic cancers.7–10 Until now, although several PTP1B inhibitors have been explored as potential anti-diabetic treatments targeting PTP1B, these agents have faced considerable challenges due to side effects.11 Hence, it is essential to discover new therapeutic agents from natural resources to minimize the adverse effects associated with long-term treatment.
Flavonoids are the most abundant natural products, attracting significant interest from researchers due to their promising health benefits and nutritional value. Hence, incorporating flavonoid-rich foods into the daily diet is essential to promoting health conditions and eliminating the risk of ailments.12 Among their numerous biological effects, flavonoids have been extensively studied for their anti-diabetic properties. They contribute to diabetes management by regulating carbohydrate digestion, enhancing insulin signaling and secretion, and promoting glucose uptake.13 Thus, exploring flavonoids as a potential therapeutic strategy for diabetes treatment is crucial.
Astragalus membranaceus Bunge (Fabaceae) has been known for its diverse biological properties, such as immunomodulation, antioxidant properties, antitumor effects, anti-diabetic activity, antiviral actions, hepatoprotection, anti-inflammatory effects, anti-atherosclerotic benefits, hematopoiesis, and neuroprotection.14 Until now, many studies have been undertaken to evaluate the anti-diabetic effects of phytoconstituents, particularly focusing on several target proteins, including α-amylase, α-glucosidase, and PTP1B.15–17 Among bioactive secondary metabolisms, astragalus polysaccharides, a key bioactive component derived from the roots of A. membranaceus, have emerged as a promising candidate for reducing PTP1B expression.18 In addition, astragaloside IV, a well-known bioactive component of this plant, was investigated for its antidiabetic effects through the PTP1B inhibitory mechanism.19 While polysaccharides demonstrated effectiveness against PTP1B, the potential biological effects of flavonoid components remain unexplored. Although the flavonoid fraction from the roots of A. membranaceus showed significant anti-diabetic activity,20,21 the number of studies focusing on the PTP1B inhibitory effects of flavonoids from the roots of A. membranaceus remains limited, leading to gaps in understanding the role of flavonoids from this plant in diabetes treatment, highlighting the need for further investigations.
Therefore, this study aimed to extract the flavonoids from the roots of A. membranaceus, investigate their PTP1B inhibitory activity, identify the structure-activity relationships responsible for PTP1B inhibition, and simultaneously elucidate the action mechanism of the most potent inhibitors against PTP1B. As a result, we successfully isolated 24 flavonoids from this plant, categorized into isoflavones (1–10), isoflavanes (11–16), flavanone (17), chalcone (18), and pterocarpans (19–24). The structures of the isolated compounds were determined using NMR spectroscopy. Additionally, an in vitro assay was performed to evaluate the PTP1B inhibitory activity and identify the inhibition mode of the most potent inhibitors. Molecular docking was employed to study the interactions between the most promising ligands and PTP1B.
Experimental
General experimental procedures – Column chromatography (CC) was utilized for the separation process, employing silica gel 60 S (0.060–0.200 mm), silica gel 60 (0.040–0.063 mm), LiChroprep RP-18 (40–63 μm), and MCI gel CHP20P (75–100 μm), provided by Merck (Darmstadt, Germany). Chromatographic separation via HPLC was executed on a Waters Alliance system (Massachusetts, USA) equipped with a 1525 binary pump, a 2998 PDA detector, and a YMC Pack ODS column (20 × 250 mm, 5 μm). Precoated TLC plates with silica gel 60 F254 and RP-18 F254s were obtained from Merck (Darmstadt, Germany). The spots were examined for UV absorbance at 254 and 365 nm, treated with 10% sulfuric acid spray, and heated at 100°C for 1 minute. NMR spectra acquisition was carried out with a Bruker Avance 500 MHz spectrometer (Bruker, Karlsruhe, Germany). Optical rotations were acquired using a JASCO P2000 polarimeter (JASCO, Tokyo, Japan). All solvents required for these experiments were obtained from Fisher Scientific Korea Ltd. (Seoul, Korea).
Plant materials – The roots of Astragalus membranaceus Bunge were obtained from a traditional market in Jeonju, Korea, in March of 2023 and authenticated by Professor Byung Sun Min at Daegu Catholic University. A documented specimen (26A-AMR) was archived at the Pharmacognosy Laboratory in the College of Pharmacy, Kyungpook National University, Korea.
Extraction and isolation – The dried roots (14.8 kg) of A. membranaceus were extracted three times using methanol (MeOH, 15 L for 4 hours each). The MeOH extract (2.1 kg) was dissolved in H2O and then partitioned sequentially with n-hexane, methylene chloride (CH2Cl2), ethyl acetate (EtOAc), and n-butanol (BuOH). The CH2Cl2 and EtOAc layers (67.6 g) were combined and subfractioned through vacuum liquid chromatography (VLC) (CH2Cl2: MeOH: H2O; 20:1:0 → 2:1:0.2, v/v/v) to obtain 4 subfractions (1A–1D). The fraction 1A was separated into 4 subfractions (2A–2D) using CC, silica gel (CH2Cl2:MeOH:H2O; 20:1:0 → 2:1:0.2, v/v/v). From fraction 2B, 9 subfractions (3A–3I) were obtained using silica gel column chromatography (CC) RP-18 (MeOH:H2O; 1:2 → 1:0, v/v). The fraction 3H was then loaded on CC, silica gel (CH2Cl2:acetone; 20:1 → 0:1, v/v) to obtain compound 1. The fractions 3D and 3E were combined and analyzed in CC, silica gel (CH2Cl2:acetone; 25:1 → 0:1, v/v) to obtain compound 20. The fraction 3G was then purified through CC, silica gel (CH2Cl2:acetone; 20:1 → 0:1, v/v) to gain compound 4, while fraction 3F was applied on CC, silica gel (Hexane:EtOAc; 2:1 → 0:1, v/v) to obtain compounds 11 and 16. Fraction 2C was separated into 12 subfractions (4A–4L) by using CC, silica gel (CH2Cl2:EtOAc; 20:1 → 0:1, v/v). Compound 2 was isolated from subfraction 4L by silica gel CC (CH2Cl2:acetone; 10:1, v/v), and compound 17 was purified from subfraction 4J through CC, RP-18 (MeOH:H2O; 1:2 → 1:0, v/v), while compounds 3, 5 and 18 were isolated from subfraction 4G in the same isolating condition. The mixture of fractions 4C and 4D was separated into 11 subfractions (5A–5J) through CC, RP-18 (acetone:H2O; 1:2 → 1:0, v/v). Subfaction 5G was determined as compound 15, while compounds 19 and 21 were isolated from subfraction 5E. Fraction 1B was fractionated into 6 subfractions (6A–6E) by using MCI gel (MeOH:H2O; 1:2 → 1:0, v/v). Compounds 23 was crystalized in MeOH from fraction 6B. From fraction 6D, 6 subfractions (8A–8F) were obtained through eluting on CC, RP-18 (MeOH:H2O; 3:2, v/v). From fraction 8D, compound 24 was isolated using silica gel CC (CH2Cl2:MeOH:H2O; 20:1:0 → 10:1:0.1, v/v/v). Fraction 8C was divided into 3 subfractions (9A–9C) by eluting on RP-18 CC (acetone:H2O; 6:5 → 1:0, v/v). Fraction 9B was then separated into 7 subfractions (10A–10G) through silica gel CC (CH2Cl2:MeOH:H2O; 25:1:0 → 8:1:0.05, v/v/v). Compounds 7 and 13 were isolated from fraction 10F by using silica gel CC (CH2Cl2:EtOAc; 1:10 → 1:20, v/v). Fraction 6C was continuously separated into 10 subfractions (11A–11J) by eluting on RP-18 CC (MeOH:H2O; 1:2 → 1:0, v/v), in which fraction 11G was determined as compound 10. Fractions 1C and 1D were combined and analyzed on silica gel CC (CH2Cl2:MeOH; 2:1 → 0:1, v/v) to obtain 6 subfractions (12A–12F). From the mixture of fraction 12B and 12C, compound 12 was crystalized in MeOH, and the remaining part was loaded on RP-18 CC (acetone:H2O; 1:3 → 1:0, v/v) to separate into 8 subfractions (13A–13H). From fraction 13B, compounds 9 and 22 were isolated through RP-18 CC (MeOH:H2O; 1:3 → 1:0, v/v). Fraction 12D was loaded on silica gel CC (CH2Cl2:MeOH; 15:1, v/v) to separate into 3 subfractions (15A–15C). Fraction 15C was fractionated into 4 subfractions (16A–16D) through RP-18 CC (MeOH:H2O; 1:2 → 1:0, v/v). Compound 8 was obtained from fraction 16B by eluting on RP-18 CC (MeOH:H2O; 2:5, v/v).
The n-BuOH layer (139.7 g) was separated into 6 fractions (18A–18F) through silica gel CC (CH2Cl2:MeOH; 1:0 → 1:1, v/v). From fraction 18C, 6 subfractions (19A–19F) were obtained by eluting on RP-18 CC (MeOH:H2O; 2:3 → 1:0, v/v), then fraction 19D was loaded on silica gel CC (CH2Cl2:MeOH; 10:1, v/v) to purify compound 6. Fraction 18F was loaded on silica gel CC (EtOAc:MeOH:H2O; 3:1:0 → 2:1:0.2, v/v/v) to separate into 3 fractions (25A– 25C). From fraction 25B, 9 subfractions (26A–26I) were obtained by using MCI gel CC (acetone:H2O; 1:3 → 1.1, v/v). Compound 14 were isolated from fraction 26B through RP-18 CC (MeOH:H2O 1:2 → 1:1, v/v).
Formonentin (1) – White amorphous powder; 1H-NMR (500 MHz, DMSO-d6): δ 8.33 (1H, s, H-2), 8.00 (1H, d, J = 8.76 Hz, H-5), 6.93 (1H, dd, J = 2.2, 8.77 Hz, H-6), 6.87 (1H, d, J = 2.20 Hz, H-8), 7.50 (1H, d, J = 8.78 Hz, H-2ʹ), 6.98, (1H, d, J = 8.81 Hz, H-3ʹ), 6.98, (1H, d, J = 8.81 Hz, H-5ʹ), 7.50 (1H, d, J = 8.78 Hz, H-6ʹ), 3.78 (3H, s, OCH3); 13C-NMR (125 MHz, DMSO-d6): δ 153.1 (C-2), 123.1 (C-3), 174.6 (C-4), 116.6 (C-4a), 127.3 (C-5), 115.2 (C-6), 162.6 (C-7), 102.1 (C-8), 157.4 (C-8a), 124.2 (C-1ʹ), 130.1 (C-2ʹ and C-6ʹ), 113.6 (C-3ʹand C-5ʹ), 158.9 (C-4ʹ), 55.1 (OCH3).
Calycosin (2) – White amorphous powder; 1H-NMR (500 MHz, CD3OD): δ 8.12 (1H, s, H-2), 8.06 (1H, d, J = 8.83 Hz, H-5), 6.94 (1H, dd, J = 2.28, 8.83 Hz, H-6), 6.85 (1H, d, J = 2.25 Hz, H-8), 7.05 (1H, s, H-2ʹ), 6.97 (2H, br s, H-5ʹ and H-6ʹ), 3.89 (3H, s, OCH3); 13C-NMR (125 MHz, CD3OD): δ 154.8 (C-2), 125.7 (C-3), 178.0 (C-4), 118.2 (C-4a), 128.5 (C-5), 116.5 (C-6), 164.7 (C-7), 103.2 (C-8), 159.7 (C-8a), 126.2 (C-1ʹ), 117.4 (C-2ʹ), 147.4 (C-3ʹ), 149.1 (C-4ʹ), 112.6 (C-5ʹ), 121.6 (C-6ʹ), 56.4 (OCH3).
Pratensein (3) – Pale yellow amorphous powder; 1H-NMR (500 MHz, DMSO-d6): δ 8.31 (1H, s, H-2), 6.21 (1H, d, J = 2.06 Hz, H-6), 6.37 (1H, d, J = 2.04 Hz, H-8), 7.03 (1H, d, J = 1.99 Hz, H-2ʹ), 6.97 (1H, d, J = 8.38 Hz, H-5ʹ), 6.93 (1H, dd, J = 2.01, 8.29 Hz, H-6ʹ), 3.79 (3H, s, OCH3); 13C-NMR (125 MHz, DMSO-d6): δ 154.1 (C-2), 123.3 (C-3), 180.0 (C-4), 104.3 (C-4a), 161.9 (C-5), 99.0 (C-6), 164.7 (C-7), 93.7 (C-8), 157.5 (C-8a), 122.1 (C-1ʹ), 116.3 (C-2ʹ), 146.1 (C-3ʹ), 147.7 (C-4ʹ), 112.0 (C-5ʹ), 119.7 (C-6ʹ), 55.6 (OCH3).
Afromorsin (4) – Yellow amorphous powder; 1H-NMR (500 MHz, DMSO-d6): δ 8.29 (1H, s, H-2), 7.39 (1H, s, H-5), 6.95 (1H, s, H-8), 7.52 (2H, d, J = 8.81 Hz, H-2ʹ and H-6ʹ), 6.98 (2H, d, J = 8.84 Hz, H-3ʹ and H-5ʹ), 3.86 (3H, s, 6-OCH3), 3.78 (3H, s, 4ʹ-OCH3); 13C-NMR (125 MHz, DMSO-d6): δ 152.4 (C-2), 122.3 (C-3), 174.1 (C-4), 115.1 (C-4a), 104.2 (C-5), 147.3 (C-6), 152.5 (C-7), 102.5 (C-8), 152.0 (C-8a), 124.5 (C-1ʹ), 129.9 (C-2 and C-6ʹ), 113.4 (C-3ʹ and C-5ʹ), 158.7 (C-4ʹ), 55.7 (6-OCH3), 55.1 (4ʹ-OCH3).
Odoratin (5) – White amorphous powder; 1H-NMR (500 MHz, CD3OD): δ 8.15 (1H, s, H-2), 7.57 (1H, s, H-5), 6.93 (1H, s, H-8), 7.07 (1H, s, H-2ʹ), 6.99 (2H, overlapped, H-5ʹ and H-6ʹ), 3.96 (3H, s, 6-OCH3), 3.98 (3H, s, 4ʹ-OCH3); 13C-NMR (125 MHz, CD3OD): δ 154.6 (C-2), 125.3 (C-3), 177.7 (C-4), 117.8 (C-4a), 105.4 (C-5), 148.8 (C-6), 154.6 (C-7), 103.7 (C-8), 154.5 (C-8a), 126.4 (C-1ʹ), 117.5 (C-2ʹ), 147.4 (C-3ʹ), 149.1 (C-4ʹ), 112.7 (C-5ʹ), 121.6 (C-6ʹ), 56.6 (6-OCH3), 56.4 (4ʹ-OCH3).
Ononin (6) – White amorphous powder; 1H-NMR (500 MHz, DMSO-d6): δ 8.43 (1H, s, H-2), 8.06 (1H, d, J = 8.87 Hz, H-5), 7.15 (1H, dd, J = 2.24, 8.88 Hz, H-6), 7.24 (1H, d, J = 2.20 Hz, H-8), 7.53 (2H, d, J = 8.73 Hz, H-2ʹ and H-6ʹ), 7.00 (2H, d, J = 8.77 Hz, H-3ʹ and H-5ʹ), 5.11 (1H, d, J = 7.22 Hz, H-1ʹʹ), 3.31 (1H, m, H-2ʹʹ), 3.47 (2H, overlapped, H-3ʹʹ and H-6ʹʹα), 3.18 (1H, m, H-4ʹʹ), 3.32 (1H, m, H-5ʹʹ), 3.73 (1H, m, H-6ʹʹβ), 3.79 (3H, s, 4ʹ- OCH3); 13C-NMR (125 MHz, DMSO-d6): δ 153.6 (C-2), 124.0 (C-3), 174.7 (C-4), 118.5 (C-4a), 127.0 (C-5), 115.6 (C-6), 161.5 (C-7), 103.4 (C-8), 157.1 (C-8a), 123.4 (C-1ʹ), 130.1 (C-2ʹ and C-6ʹ), 113.6 (C-3ʹ and C-5ʹ), 159.0 (C-4ʹ), 100.0 (C-1ʹʹ), 73.1 (C-2ʹʹ), 77.2 (C-3ʹʹ), 69.6 (C-4ʹʹ), 76.5 (C-5ʹʹ), 60.6 (C-6ʹʹ), 55.2 (4ʹ-OCH3).
6′′-O-Acetylononin (7) – Pale yellow amorphous powder; 1H-NMR (500 MHz, CD3OD): δ 8.22 (1H, s, H-2), 8.15 (1H, d, J = 8.86 Hz, H-5), 7.20 (1H, dd, J = 2.21, 8.96 Hz, H-6), 7.22 (1H, d, J = 2.10 Hz, H-8), 7.49 (2H, d, J = 8.74 Hz, H-2ʹ and H-6ʹ), 6.99 (2H, m, H-3ʹ and H-5ʹ), 5.12 (1H, m, H-1ʹʹ), 3.55 (2H, overlapped, H-2ʹʹ and H-3ʹʹ), 3.43 (1H, m, H-4ʹʹ), 3.79 (1H, m, H-5ʹʹ), 4.48 (1H, m, H-6ʹʹα), 4.25 (1H, m, H-6ʹʹβ), 3.84 (3H, s, COCH3), 2.10 (3H, s, 4ʹ-OCH3); 13C-NMR (125 MHz, CD3OD): δ 155.2 (C-2), 126.0 (C-3), 177.9 (C-4), 120.3 (C-4a), 128.3 (C-5), 117.1 (C-6), 163.3 (C-7), 105.0 (C-8), 159.1 (C-8a), 125.3 (C-1ʹ), 131.3 (C-2ʹ and C-6ʹ), 114.9 (C-3ʹ and C-5ʹ), 161.2 (C-4ʹ), 101.7 (C-1ʹʹ), 74.7 (C-2ʹʹ), 77.7 (C-3ʹʹ), 71.5 (C-4ʹʹ), 75.6 (C-5ʹʹ), 64.7 (C-6ʹʹ), 172.6 (COCH3), 20.7 (COCH3), 55.7 (4ʹ-OCH3).
Calycosin 7-O-β-D-glucoside (8) – White amorphous powder; 1H-NMR (500 MHz, DMSO-d6): δ 8.36 (1H, s, H-2), 8.06 (1H, d, J = 8.75 Hz, H-5), 7.15 (1H, dd, J = 2.50, 8.49 Hz, H-6) 7.23 (1H, d, J = 2.5 Hz, H-8), 7.09 (1H, m, H-2ʹ), 6.96 (2H, overlapped, H-5ʹ and H-6ʹ), 5.12 (1H, br d, H-1ʹʹ), 3.35 (2H, m, H-2ʹʹ and H-3ʹʹ), 3.22 (1H, m, H-4ʹʹ), 3.49 (2H, m, H-5ʹʹ and H-6ʹʹβ), 3.77 (1H, m, H-6ʹʹα), 3.79 (3H, s, 4ʹ-OCH3); 13C-NMR (125 MHz, DMSO-d6): δ 153.6 (C-2), 123.7 (C-3), 174.8 (C-4), 118.6 (C-4a), 127.1 (C-5), 115.7 (C-6), 161.5 (C-7), 103.5 (C-8), 157.1 (C-8a), 124.4 (C-1ʹ), 116.5 (C-2ʹ), 146.1 (C-3ʹ), 147.7 (C-4ʹ), 112.0 (C-5ʹ), 119.8 (C-6ʹ), 100.1 (C-1ʹʹ), 73.2 (C-2ʹʹ), 76.6 (C-3ʹʹ) 69.7 (C-4ʹʹ), 77.3 (C-5ʹʹ), 60.8 (C-6ʹʹ), 55.7 (4ʹ-OCH3).
Calycosin 7-O-β-D-glucoside-6′′-O-acetate (9) – Yellow amorphous powder; 1H NMR (500 MHz, CD3OD): δ 8.18 (1H, s, H-2), 8.17 (1H, d, J = 8.85 Hz, H-5), 7.23 (1H, m, H-6), 7.25 (1H, br d, H-8), 7.09 (1H, br s, H-2ʹ), 7.02 (2H, br s, H-5ʹ and H-6ʹ), 5.13 (1H, m, H-1ʹʹ), 3.56 (2H, m, H-2ʹʹ and H-3ʹʹ), 3.44 (1H, m, H-4ʹʹ), 3.80 (1H, m, H-5ʹʹ), 4.50 (1H, m, H-6ʹʹα), 4.27 (1H, m, H-6ʹʹβ), 3.92 (3H, s, COCH3), 2.11 (3H, s, 4ʹ-OCH3); 13C-NMR (125 MHz, CD3OD): δ 155.2 (C-2), 125.9 (C-3), 177.9 (C-4), 120.3 (C-4a), 128.3 (C-5), 117.1 (C-6), 163.3 (C-7), 105.0 (C-8), 159.1 (C-8a), 126.1 (C-1ʹ), 117.4 (C-2ʹ), 147.4 (C-3ʹ), 149.3 (C-4ʹ), 112.7 (C-5ʹ), 121.6 (C-6ʹ), 101.7 (C-1ʹʹ), 74.7 (C-2ʹʹ), 77.7 (C-3ʹʹ), 71.5 (C-4ʹʹ) 75.6 (C-5ʹʹ), 64.7 (C-6ʹʹ), 172.7 (COCH3), 20.7 (COCH3), 56.4 (4ʹ-OCH3).
Calycosin 7-O-β-D-{6ʹʹ-[(E)-but-2-enoyl]}-glucoside (10) – Yellow amorphous powder; 1H-NMR (500 MHz, DMSOd6): δ 8.83 (1H, s, H-2), 8.04 (1H, d, J = 8.86 Hz, H-5), 7.13 (1H, dd, J = 1.77, 8.90 Hz, H-6), 7.19 (1H, d, J = 1.74 Hz, H-8), 7.07 (1H, brs, H-2ʹ), 6.96 (2H, br s, H-5ʹ and H-6ʹ), 5.18 (1H, d, J = 6.97 Hz, H-1ʹʹ), 3.36 (2H, m, H-2ʹʹ and H-5ʹʹ), 3.80 (1H, m, H-3ʹʹ), 3.23 (1H, m, H-4ʹʹ), 4.14 (1H, br d, H-6ʹʹα), 4.13 (1H, br d, H-6ʹʹβ), 5.88 (1H, d, J = 15.49 Hz, H-2ʹʹʹ), 6.88 (1H, dd, J = 6.96, 15.46 Hz, H-3ʹʹʹ), 1.83 (3H, d, J = 6.32 Hz, H-4ʹʹʹ), 3.79 (3H, s, 4ʹ-OCH3); 13C-NMR (125 MHz, DMSO-d6): δ 153.6 (C-2), 124.5 (C-3), 174.7 (C-4), 118.6 (C-4a), 127.0 (C-5), 115.6 (C-6), 161.2 (C-7), 103.4 (C-8), 157.0 (C-8a), 123.7 (C-1ʹ), 116.5 (C-2ʹ), 146.1 (C-3ʹ), 147.7 (C-4ʹ), 112.0 (C-5ʹ), 119.8 (C-6ʹ), 99.6 (C-1ʹʹ), 73.1 (C-2ʹʹ), 73.9 (C-3ʹʹ), 70.1 (C-4ʹʹ), 76.3 (C-5ʹʹ), 63.3 (C-6ʹʹ), 165.3 (C-1ʹʹʹ), 122.1 (C-2ʹʹʹ), 145.4 (C-3ʹʹʹ), 17.7 (C-4ʹʹʹ), 55.7 (4ʹ-OCH3).
Isomucronulatol (11) – White amorphous powder; [α]D25‒11.67 (c 0.02, MeOH); 1H-NMR (500 MHz, CD3OD): δ 4.25 (1H, ddd, J = 2.01, 3.38, 10.29 Hz, H-2α), 3.97 (1H, t, J = 10.1 Hz, H-2β), 3.47 (1H, m, H-3), 2.95 (1H, dd, J = 10.87, 15.48 Hz, H-4α), 2.80 (1H, ddd, J = 1.49, 5.06, 15.62 Hz, H-4β), 6.88 (1H, d, J = 8.23 Hz, H-5), 6.35 (1H, dd, J = 2.47, 8.22 Hz, H-6), 6.27 (1H, d, J = 2.44 Hz, H-8), 6.46 (1H, d, J = 8.67 Hz, H-5ʹ), 6.77 (1H, d, J = 8.65 Hz, H-6ʹ), 3.82 (6H, br s, 3ʹ-OCH3 and 4ʹ-OCH3); 13C-NMR (125 MHz, CD3OD): δ 70.9 (C-2), 33.5 (C-3), 31.2 (C-4), 114.8 (C-4a), 131.2 (C-5), 109.3 (C-6), 157.0 (C-7), 103.8 (C-8), 156.3 (C-8a), 122.4 (C-1ʹ), 149.5 (C-2ʹ), 137.5 (C-3ʹ), 153.1 (C-4ʹ), 104.4 (C-5ʹ), 122.8 (C-6ʹ), 56.2 (3ʹ-OCH3), 61.0 (4ʹ-OCH3).
Isomucronulatol 7-O-β-glucoside (12) – White, crystalline powder; [α]D25‒43.55 (c 0.02, MeOH); 1H-NMR (500 MHz, DMSO-d6): δ 4.19 (1H, ddd, J = 10.5 Hz, 3.5 Hz, 1.7 Hz, H-2α), 3.96 (1H, t, J = 10.02 Hz, H-2β), 3.36 (1H, m, H-3), 2.93 (1H, dd, J=10.6, 15.85 Hz, H-4α), 2.80 (1H, dd, J = 4.3, 15.82 Hz, H-4β), 7.00 (1H, d, J = 8.43 Hz, H-5), 6.55 (1H, dd, J = 2.36, 8.33 Hz, H-6), 6.47 (1H, d, J = 2.46 Hz, H-8), 6.46 (1H, d, J = 8.55 Hz, H-5ʹ), 6.78 (1H, d, J = 8.66 Hz, H-6ʹ), 4.77 (1H, d, J = 7.63 Hz, H-1ʹʹ), 3.20 (1H, m, H-2ʹʹ), 3.28 (2H, m, H-3ʹʹ and H-5ʹʹ), 3.14 (1H, m, H-4ʹʹ), 3.69 (1H, overlapped, H-6ʹʹα), 3.46 (1H, m, H-6ʹʹβ), 3.68 (3H, s, 3ʹ-OCH3), 3.74 (3H, s, 4ʹ-OCH3); 13C-NMR (125 MHz, DMSO-d6): δ 69.2 (C-2), 31.3 (C-3), 29.7 (C-4), 115.8 (C-4a), 130.0 (C-5), 108.8 (C-6), 156.7 (C-7), 103.8 (C-8), 154.5 (C-8a), 120.8 (C-1ʹ), 148.1 (C-2ʹ), 136.2 (C-3ʹ), 151.7 (C-4ʹ), 103.2 (C-5ʹ), 121.4 (C-6ʹ), 100.7 (C-1ʹʹ), 73.2 (C-2ʹʹ), 76.6 (C-3ʹʹ), 69.8 (C-4ʹʹ), 77.0 (C-5ʹʹ), 60.7 (C-6ʹʹ), 60.2 (3ʹ-OCH3), 55.6 (4ʹ-OCH3).
2′-Hydroxy-3′,4′-dimethoxyisoflavan 7-O-β-D-glucoside 6′′-O-acetate (13) – White amorphous powder; [α]D25‒36.48 (c 0.04, MeOH); 1H-NMR (500 MHz, CD3OD): δ 4.29 (1H, m, H-2α), 4.03 (1H, t, J = 10.04 Hz, H-2β), 3.48 (1H, overlapped, H-3), 3.02 (1H, dd, J = 10.59, 15.84 Hz, H-4α), 2.88 (1H, m, H-4β), 7.00 (1H, d, J = 8.35 Hz, H-5), 6.61 (1H, dd, J = 2.48, 8.34 Hz, H-6), 6.57 (1H, d, J = 2.45 Hz, H-8), 6.48 (1H, d, J = 8.70 Hz, H-5ʹ), 6.80 (1H, d, J = 8.66 Hz, H-6ʹ), 4.84 (1H, d, J = 7.46 Hz, H-1ʹʹ), 3.47 (1H, m, H-2ʹʹ), 3.65 (1H, m, H-3ʹʹ), 3.38 (1H, m, H-4ʹʹ), 3.48 (1H, m, H-5ʹʹ), 4.42 (1H, dd, J = 2.20, 11.81 Hz, H-6ʹʹα), 4.19 (1H, m, H-6ʹʹβ), 2.08 (3H, s, COCH3), 3.82 (3H, s, 3ʹ-OCH3), 3.83 (3H, s, 4ʹ-OCH3); 13C-NMR (125 MHz, CD3OD): δ 71.0 (C-2), 33.4 (C-3), 31.3 (C-4), 117.9 (C-4a), 131.1 (C-5), 110.4 (C-6), 158.2 (C-7), 105.6 (C-8), 156.3 (C-8a), 122.3 (C-1ʹ), 149.5 (C-2ʹ), 137.6 (C-3ʹ), 153.2 (C-4ʹ), 104.4 (C-5ʹ), 122.8 (C-6ʹ), 102.4 (C-1ʹʹ), 74.9 (C-2ʹʹ), 75.3 (C-3ʹʹ), 71.8 (C-4ʹʹ), 77.9 (C-5ʹʹ), 64.8 (C-6ʹʹ), 172.8 (COCH3), 20.7 (COCH3), 61.1 (3ʹ-OCH3), 56.3 (4ʹ-OCH3).
Isomucronulatol 2′,7-diglucoside (14) – White amorphous powder; [α]D25‒21.78 (c 0.04, MeOH); 1H-NMR (500 MHz, CD3OD): δ 4.4 (1H, m, H-2α), 3.91 (1H, m, H-2β), 3.82 (1H, m, H-3), 2.91 (1H, dd, J = 11.05, 15.86 Hz, H-4α), 2.82 (1H, dd, J = 4.66, 15.45 Hz, H-4β), 6.99 (1H, d, J = 8.39 Hz, H-5), 6.65 (1H, dd, J = 2.4, 8.36 Hz, H-6), 6.60 (1H, d, J = 2.44 Hz, H-8), 6.83 (1H, d, J = 8.78 Hz, H-5ʹ), 6.91 (1H, d, J = 8.74 Hz, H-6ʹ), 4.89 (1H, m, H-1ʹʹ), 3.46 (2H, m, H-2ʹʹ and H-3ʹʹ), 3.44 (2H, m, H-4ʹʹ and H-3ʹʹʹ), 3.45 (1H, m, H-5ʹʹ), 3.93 (1H, m, H-6ʹʹα), 3.70 (1H, m, H-6ʹʹβ), 5.06 (1H, m, H-1ʹʹʹ), 3.47 (1H, m, H-2ʹʹʹ), 3.38 (1H, m, H-4ʹʹʹ), 3.35 (1H, m, H-5ʹʹʹ), 3.92 (1H, m, H-6ʹʹʹα), 3.72 (1H, m, H-6ʹʹʹβ), 3.87 (3H, s, 3ʹ-OCH3), 3.86 (3H, s, 4ʹ-OCH3); 13C-NMR (125 MHz, CD3OD): δ 71.7 (C-2), 32.1 (C-3), 32.5 (C-4), 118.0 (C-4a). 131.2 (C-5), 110.3 (C-6), 158.4 (C-7), 105.7 (C-8), 156.3 (C-8a), 130.2 (C-1ʹ), 149.2 (C-2ʹ), 143.0 (C-3ʹ), 153.9 (C-4ʹ), 109.9 (C-5ʹ), 122.9 (C-6ʹ), 102.5 (C-1ʹʹ), 75.1 (C-2ʹʹ), 77.9 (C-3ʹʹ), 71.3 (C-4ʹʹ), 78.1 (C-5ʹʹ), 62.8 (C-6ʹʹ), 105.1 (C-1ʹʹʹ), 74.9 (C-2ʹʹʹ), 77.8 (C-3ʹʹʹ), 71.6 (C-4ʹʹʹ), 78.3 (C-5ʹʹʹ), 62.4 (C-6ʹʹʹ), 61.6 (3ʹ-OCH3), 56.5 (4ʹ-OCH3).
Pendulone (15) – Reddish crystalline powder; [α]D25‒19.05 (c 0.02, MeOH); 1H NMR (500 MHz, CD3OD): δ 4.24 (1H, ddd, J = 1.3, 3.04, 10.64, H-2α), 3.98 (1H, m, H-2β), 3.37 (1H, d, J = 7.23 Hz, H-3), 2.95 (1H, dd, J = 5.61, 15.8 Hz, H-4α), 2.74 (1H, dd, J = 7.93, 15.84 Hz, H-4β), 6.89 (1H, d, J = 8.32 Hz, H-5), 6.36 (1H, dd, J = 2.46, 8.26 Hz, H-6), 6.25 (1H, d, J = 2.44 Hz, H-8), 6.40 (1H, s, H-6ʹ), 4.01 (6H, br s, 3ʹ-OCH3 and 4ʹ-OCH3); 13CNMR (125 MHz, CD3OD): δ 69.4 (C-2), 32.4 (C-3), 30.7 (C-4), 112.8 (C-4a), 131.2 (C-5), 109.7 (C-6), 158.0 (C-7), 103.9 (C-8), 156.0 (C-8a), 148.2 (C-1ʹ), 184.8 (C-2ʹ), 146.7 (C-3ʹ), 146.1 (C-4ʹ), 184.4 (C-5ʹ), 131.9 (C-6ʹ), 61.7 (3ʹ-OCH3), 61.6 (4ʹ-OCH3).
4,7,2ʹ-Trihydroxy-4ʹ-methoxyisoflavanol (16) – Yellow powder; [α]D25‒170.01 (c 0.02, MeOH); 1H-NMR (500 MHz, CD3OD) δ 4.24 (1H, dd, J = 2.83, 8.59 Hz, H-2α), 3.54 (1H, m, H-2β), 4.56 (1H, m, H-3), 5.49 (1H, d, J = 6.36 Hz, H-4), 7.31 (1H, d, J = 8.43 Hz, H-5), 6.52 (1H, dd, J = 2.43, 8.40 Hz, H-6), 6.33 (1H, d, J = 2.41 Hz, H-8), 6.41 (1H, d, J = 2.26 Hz, H-3ʹ), 6.46 (1H, dd, J = 2.28, 8.18 Hz, H-5ʹ), 7.19 (1H, d, J = 8.19 Hz, H-6ʹ); 13C-NMR (125 MHz, CD3OD): δ 67.5 (C-2), 40.9 (C-3), 80.1 (C-4), 112.9 (C-4a), 133.1 (C-5), 110.7 (C-6), 160.1 (C-7), 104.9 (C-8), 158.1 (C-8a), 120.8 (C-1ʹ), 162.1 (C-2ʹ), 97.6 (C-3ʹ), 162.6 (C-4ʹ), 107.2 (C-5ʹ), 125.9 (C-6ʹ).
(2S)-Liquiritigenin (17) – Pale yellow powder; [α]D25‒18.30 (c 0.08, MeOH); 1H-NMR (500 MHz, CD3OD) δ 5.41 (1H, dd, J = 2.88, 13.6 Hz, H-2), 3.08 (1H, dd, J = 13.07, 16.91 Hz, H-3α), 2.72 (1H, dd, J = 2.96, 16.91 Hz, H-3β), 7.76 (1H, d, J = 8.76 Hz, H-5), 6.55 (1H, dd, J = 2.27, 8.72 Hz, H-6), 6.38 (1H, d, J = 2.24 Hz, H-8), 7.35 (1H, d, J = 8.47 Hz, H-2ʹ), 6.84 (1H, d, J = 8.63 Hz, H-3ʹ), 6.83 (1H, d, J = 8.63 Hz, H-5ʹ), 7.35 (1H, d, J = 8.47 Hz, H-6ʹ); 13C-NMR (125 MHz, CD3OD): δ 81.1 (C-2), 44.6 (C-3), 193.5 (C-4), 115.0 (C-4a), 129.9 (C-5), 111.7 (C-6), 166.8 (C-7), 103.8 (C-8), 165.6 (C-8a), 131.4 (C-1ʹ), 129.0 (C-2ʹ), 116.3 (C-3ʹ), 158.9 (C-4ʹ), 116.3 (C-5ʹ), 129.0 (C-6ʹ).
Isoliquiritigenin (18) – Yellow crystalline powder; 1H-NMR (500 MHz, CD3OD) δ 7.64 (2H, d, J = 8.91 Hz, H-2 and H-6), 6.86 (2H, d, J = 8.60 Hz, H-3 and H-5), 6.31 (1H, d, J = 2.37 Hz, H-3ʹ), 6.43 (1H, dd, J = 2.37, 8.87 Hz, H-5ʹ), 7.99 (1H, d, J = 8.91 Hz, H-6ʹ), 7.64 (1H, overlapped, CH-α), 7.80 (1H, d, J = 15.35 Hz, CH-β); 13C-NMR (125 MHz, CD3OD): δ 127.9 (C-1), 131.8 (C-2 and C-6), 116.9 (C-3 and C-5), 161.6 (C-4), 114.7 (C-1ʹ), 166.5 (C-2ʹ), 103.9 (C-3ʹ), 167.5 (C-4ʹ), 109.2 (C-5ʹ), 133.4 (C-6ʹ), 118.4 (CH-α), 145.6 (CH-β), 193.5 (C=O).
Vesticarpan (19) – Pale yellow powder; [α]D25‒155.69 (c 0.02, MeOH); 1H-NMR (500 MHz, CD3OD) δ 7.37 (1H, d, J = 8.43 Hz, H-1), 6.52 (1H, dd, J = 2.44, 8.40 Hz, H-2), 6.33 (1H, d, J = 2.40 Hz, H-4), 4.24 (1H, dd, J = 4.33, 10.40 Hz, H-6α), 3.57 (1H, t, J = 10.40 Hz, H-6β), 3.52 (1H, ddd, J = 4.33, 6.20, 10.40 Hz, H-6a), 6.77 (1H, d, J = 8.19 Hz, H-7), 6.52 (1H, d, J = 8.13 Hz, H-8), 5.50 (1H, d, J = 6.24 Hz, H-11a), 3.84 (3H, s, 9-OCH3); 13CNMR (125 MHz, CD3OD): δ 133.4 (C-1), 110.7 (C-2), 160.1 (C-3), 104.0 (C-4), 158.0 (C-4a), 67.5 (C-6), 41.5 (C-6a), 122.9 (C-6b), 115.6 (C-7), 105.8 (C-8), 150.5 (C-9), 132.5 (C-10), 148.6 (C-10a), 80.4 (C-11a), 112.8 (C-11b), 56.7 (9-OCH3).
Methylnissolin (20) – White, amorphous powder; [α]D25‒177.97 (c 0.04, MeOH); 1H-NMR (500 MHz, CD3OD) δ 6.97 (1H, d, J = 8.16 Hz, H-1), 6.52 (1H, m, H-2), 6.33 (1H, d, J = 2.40 Hz, H-4), 4.24 (1H, dd, J = 4.01, 10.30 Hz, H-6α), 3.60 (1H, t, J = 10.30 Hz, H-6β), 3.54 (1H, ddd, J = 4.01, 6.31, 10.30 Hz, H-6a), 6.97 (1H, d, J = 8.16 Hz, H-7), 6.54 (1H, d, J = 8.24 Hz, H-8), 5.53 (1H, d, J = 6.52 Hz, H-11a), 3.82 (3H, s, 9-OCH3), 3.85 (3H, s, 10-OCH3); 13C-NMR (125 MHz, CD3OD): δ 133.3 (C-1), 110.7 (C-2), 160.2 (C-3), 104.1 (C-4), 158.0 (C-4a), 67.3 (C-6), 41.2 (C-6a), 123.5 (C-6b), 119.8 (C-7), 106.2 (C-8), 154.4 (C-9), 135.7 (C-10), 152.6 (C-10a), 80.7 (C-11a), 112.7 (C-11b), 56.8 (9-OCH3), 60.9 (10-OCH3).
(6aR,11aR)-3,8-Dihydroxy-9,10-dimethoxypterocarpan (21) – Yellow powder; [α]D25‒93.49 (c 0.02, MeOH); 1H-NMR (500 MHz, CD3OD) δ 7.33 (1H, d, J = 8.42 Hz, H-1), 6.51 (1H, dd, J = 8.40, 2.42 Hz, H-2), 6.33 (1H, d, J = 2.39 Hz, H-4), 4.23 (1H, dd, J = 4.95, 10.70 Hz, H-6α), 3.62 (1H, t, J = 10.70 Hz, H-6β), 3.48 (1H, ddd, J = 10.70, 6.80, 4.95 Hz, H-6a), 6.58 (1H, s, H-7), 5.45 (1H, d, J = 6.85 Hz, H-11a), 3.81 (3H, s, 9-OCH3), 3.93 (3H, s, 10-OCH3); 13C-NMR (125 MHz, CD3OD): δ 133.2 (C-1), 110.7 (C-2), 160.1 (C-3), 104.0 (C-4), 158.0 (C-4a), 67.2 (C-6), 41.8 (C-6a), 124.8 (C-6b), 107.1 (C-7), 144.9 (C-8), 141.2 (C-9), 139.5 (C-10), 145.9 (C-10a), 79.8 (C-11a), 112.9 (C-11b), 61.5 (9-OCH3), 60.8 (10-OCH3).
Licoagroside D (22) – Yellow amorphous powder; [α]D25‒191.81 (c 0.02, MeOH); 1H-NMR (500 MHz, CD3OD): δ 7.45 (1H, d, J = 8.63 Hz, H-1), 6.79 (1H, dd, J = 2.45, 8.55 Hz, H-2), 6.64 (1H, d, J = 2.42 Hz, H-4), 4.24 (1H, dd, J = 3.96, 10.20 Hz, H-6α), 3.55 (1H, t, J = 10.20 Hz, H-6β), 3.54 (1H, ddd, J = 10.20, 6.20, 3.96 Hz, H-6a), 7.73 (1H, d, J = 8.18 Hz, H-7), 6.49 (1H, d, J = 8.18 Hz, H-8), 5.48 (1H, d, J = 6.28 Hz, H-11a), 3.80 (3H, s, 9-OCH3), 4.90 (1H, d, J = 7.41 Hz, H-1ʹ), 3.46 (2H, overlapped, H-2ʹ and H-3ʹ), 3.42 (2H, overlapped, H-4ʹ and H-5ʹ), 3.88 (1H, dd, J = 2.11, 12.11 Hz, H-6ʹα), 3.70 (1H, dd, J = 5.32, 12.12 Hz, H-6ʹβ); 13C-NMR (125 MHz, CD3OD): δ 133.3 (C-1), 111.6 (C-2), 160.2 (C-3), 105.7 (C-4), 158.0 (C-4a), 67.5 (C-6), 41.4 (C-6a), 122.6 (C-6b), 115.6 (C-7), 105.9 (C-8), 150.4 (C-9), 132.4 (C-10), 148.2 (C-10a), 80.0 (C-11a), 115.7 (C-11b), 57.0 (9-OCH3), 102.0 (C-1ʹ), 74.8 (C-2ʹ), 77.9 (C-3ʹ), 71.3 (C-4ʹ), 78.1 (C-5ʹ), 62.4 (C-6ʹ).
9-O-Methylnissolin 3-O-glucoside (23) – White amorphous powder; [α]D25‒133.55 (c 0.02, MeOH); 1H-NMR (500 MHz, DMSO-d6): δ 7.42 (1H, d, J = 8.63 Hz, H-1), 6.73 (1H, dd, J = 2.43, 8.53 Hz, H-2), 6.56 (1H, d, J = 2.41 Hz, H-4), 4.28 (1H, dd, J = 3.90, 10.20 Hz, H-6α), 3.67 (1H, t, J = 10.20 Hz, H-6β), 3.68 (1H, ddd, J = 10.20, 6.80, 3.90 Hz, H-6a), 7.00 (1H, d, J = 8.24 Hz, H-7), 6.53 (1H, d, J = 8.26 Hz, H-8), 5.63 (1H, d, J = 6.79 Hz, H-11a), 3.73 (3H, s, 9-OCH3), 3.71 (3H, s, 10-OCH3), 4.85, (1H, d, J = 7.54 Hz, H-1ʹ), 3.22 (1H, m, H-2ʹ), 3.29 (1H, m, H-3ʹ), 3.15 (1H, m, H-4ʹ), 3.30 (1H, m, H-5ʹ), 4.28 (1H, m, H-6ʹα), 3.67 (1H, m, H-6ʹβ); 13C-NMR (125 MHz, DMSO-d6): δ 132.1 (C-1), 110.5 (C-2), 158.6 (C-3), 104.0 (C-4), 156.2 (C-4a), 65.8 (C-6), 39.2 (C-6a), 121.6 (C-6b), 118.7 (C-7), 105.1 (C-8), 152.7 (C-9), 133.4 (C-10), 151.0 (C-10a), 78.2 (C-11a), 114.1 (C-11b), 56.1 (9- OCH3), 59.9 (10-OCH3), 100.3 (C-1ʹ), 73.2 (C-2ʹ), 76.6 (C-3ʹ), 69.7 (C-4ʹ), 77.1 (C-5ʹ), 60.7 (C-6ʹ).
(-)-Methylinissolin 3-O-β-D-{6ʹʹ-[(E)-but-2-enoyl]}-glucoside (24) – Pale yellow, amorphous powder; [α]D25‒68.47 (c 0.06, MeOH); 1H-NMR (500 MHz, CD3OD): δ 7.46 (1H, d, J = 8.61 Hz, H-1), 6.80 (1H, dd, J = 2.44, 8.54 Hz, H-2), 6.67 (1H, d, J = 2.42 Hz, H-4), 4.30 (1H, dd, J = 3.99, 10.06 Hz, H-6α), 3.63 (1H, t, J = 10.06 Hz, H-6β), 3.68 (1H, ddd, J = 10.06, 6.50, 3.99 Hz, H-6a), 6.99 (1H, d, J = 8.18 Hz, H-7), 6.56 (1H, d, J = 8.22 Hz, H-8), 5.59 (1H, d, J = 6.43 Hz, H-11a), 3.83 (3H, s, 9-OCH3), 3.85 (3H, s, 10-OCH3), 4.89 (1H, m, H-1ʹ), 3.49 (2H, overlapped, H-2ʹ and H-3ʹ), 3.39 (1H, m, H-4ʹ), 3.71 (1H, m, H-5ʹ), 4.52 (1H, dd, J = 2.14, 11.89 Hz, H-6ʹα), 4.25 (1H, dd, J = 7.29, 11.91 Hz, H-6ʹβ), 5.92 (1H, dd, J = 1.74, 15.56 Hz, H-2ʹʹ), 7.04 (1H, dd, J = 6.90, 15.55 Hz, H-3ʹʹ), 1.89 (3H, dd, J = 1.70, 6.90 Hz, H-4ʹʹ); 13C-NMR (125 MHz, CD3OD): δ 133.1 (C-1), 112.0 (C-2), 160.2 (C-3), 105.8 (C-4), 157.9 (C-4a), 67.6 (C-6), 41.2 (C-6a), 123.3 (C-6b), 119.9 (C-7), 106.4 (C-8), 152.6 (C-9), 135.2 (C-10), 154.4 (C-10a), 80.3 (C-11a) 115.8 (C-11b), 56.9 (9-OCH3), 60.9 (10-OCH3), 102.1 (C-1ʹ), 74.8 (C-2ʹ), 77.9 (C-3ʹ), 71.8 (C-4ʹ), 75.5 (C-5ʹ), 64.7 (C-6ʹ), 167.9 (C-1ʹʹ), 123.2 (C-2ʹʹ), 146.8 (C-3ʹʹ), 18.2 (C-4ʹʹ).
PTP1B inhibition assay – The inhibitory activity of PTP1B was evaluated via an in vitro assay conducted on a 96-well plate. Samples were prepared at varying concent-rations (1–100 μM) by diluting them in a reaction buffer containing 50 mM citrate buffer (pH 6.0), 0.1 M NaCl, 1 mM EDTA (Sigma-Aldrich), and 1 mM dithiothreitol (DTT; Bio-Rad Laboratories, CA, USA). Next, 10 μL of the test samples were added to each well, followed by 30– 40 μL of reaction buffer and 10 μL of PTP1B enzyme (human, recombinant; NKMAX, Korea, 5 μg/mL). The mixture was incubated at 37°C for 10 minutes before adding 50 μL of 2.0 mM p-NPP (Sigma-Aldrich) substrate. Then, the reaction was mixed gently and incubated again at 37°C for 20 minutes.
The reaction was stopped by adding 10 μL of 10 M NaOH, creating an alkaline environment, resulting in the formation of the yellow p-nitrophenolate anion. This anion absorbs at 405 nm, allowing for the determination of PTP1B inhibitory activity. Ursolic acid was included as a positive control in this experiment. The inhibition percentage was calculated using the following formula:
% Inhibition = [(Δδcontrol – Δδinhibitior) / Δδ control] × 100%
Enzyme kinetic analysis – According to the PTP1B inhibition data, compounds 1, 3, and 19 emerged as the most potent inhibitors. Building on previous studies of compounds 1 and 3,22,23 this investigation aimed to explore the inhibition mechanism of compound 19. The experimental procedure followed a similar approach to the previously described PTP1B inhibition assay, with several adjustments. The test sample was prepared at various concentrations (0, 5, 7, 10, and 20 μM) and tested against different substrate concentrations (0.5, 1.0, and 2.0 mM), resulting in distinct enzymatic reactions. The Lineweaver-Burk plot was used to analyze the data, and the inhibition mode was determined based on the intersection of the trendlines. If the lines intersected on the x-axis or y-axis, it indicated a noncompetitive or competitive inhibition mode, respectively.Conversely, if the trendlines intersected in the x-y region, a mixed inhibition mode was identified. Additionally, the Dixon plot was used to calculate the inhibition constants (Ki values), which represent the binding affinity between the enzyme and the inhibitor. The figures were analyzed using Microsoft Excel 365 and visualized by Sigma plot software 15.0.
Molecular docking – Molecular docking simulations were conducted using AutoDock 4.2 software, according to our previously published protocol with a slight modification.24 Briefly, both 2D and 3D conformers of vesticarpan (19) were constructed by ChemBioOffice 16.0. The PTP1B structure and its reference catalytic inhibitor [3-({5-[(N-acetyl-3-{4-[(carboxycarbonyl) (2-carboxyphenyl)amino]-1-naphthyl}-L-alanyl)amino]pentyl}oxy)-2-naphthoic acid (compound C; PDB ID: 1NNY)] were obtained from the Protein Data Bank website (https://www.rcsb.org). A Lamarckian genetic algorithm method implemented in AutoDock 4.2 was employed. For docking calculations, Gasteiger charges were added by default, the rotatable bonds were set by the AutoDock Tools, and all torsions were allowed to rotate. The docking protocol for rigid and flexible ligand docking consisted of 15 independent Genetic Algorithms, while other parameters were used as defaults for the AutoDock Tools. The interactions of protein-ligand complexes were analyzed and visualized using PyMOL and Discovery Studio Visualizer 16.1 (Accelrys, Inc. San Diego, CA, USA).
Statistical analysis – The data were presented as means ± standard error of the mean (SEM) based on three replicates. Duncan’s test and ANOVA were applied to assess statistical significance at p < 0.05.
Results and Discussion
The structures of all isolated compounds were determined using 1D and 2D NMR spectroscopy. After the spectroscopic analysis, the obtained structures were compared with the relevant data from published studies. Therefore, the structures of 24 flavonoids were identified as formonentin (1)25 calycosin (2)26 pratensein (3)27 afromorsin (4)28 odoratin (5)29 ononin (6)30 6′′-O-Acetylononin (7)31 calycosin 7-O-β-D-glucoside (8)32 calycosin 7-O-β-Dglucoside-6′′-O-acetate (9)33 calycosin 7-O-β-D-{6ʹʹ-[(E)-but-2-enoyl]}-glucoside (10)34 isomucronulatol (11)35 isomucronulatol 7-O-β-glucoside (12)36 2′-hydroxy-3′,4′-dimethoxyisoflavan 7-O-β-D-glucoside 6′′-O-acetate (13)36 isomucronulatol 2′,7-diglucoside (14)37 pendulone (15)38 4,7,2ʹ-Trihydroxy-4ʹ-methoxyisoflavanol (16)39 (2S)-liquiritigenin (17)40 isoliquiritigenin (18)41 vesticarpan (19)41 methylnissolin (20)41 (6aR,11aR)-3,8-dihydroxy-9,10-dimethoxypterocarpan (21)41 licoagroside D (22)42 9-O-methylnissolin 3-O-glucoside (23)33 (-)-methylinissolin 3-O-β-D-{6ʹʹ-[(E)-but-2-enoyl]}-glucoside (24)33 (Fig. 1).
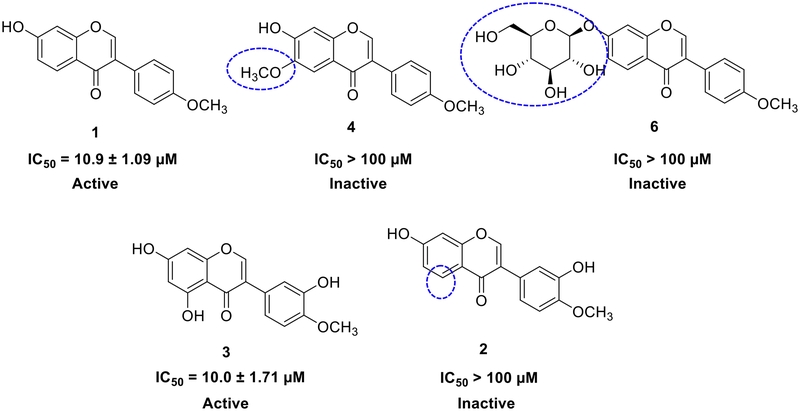
All isolated compounds were evaluated for their PTP1B inhibitory activity. As illustrated in Table 1, compared to ursolic acid (IC50 8.9 ± 1.70 μM), formonentin (1), pratensein (3), and vesticarpan (19) standed out as the promissing PTP1B inhibitors with IC50 values of 10.9 ± 1.09 μM, 10.0 ± 1.71 μM, and 10.3 ± 1.31 μM, respectively.
According to the structure-activity relationship among isoflavones (Fig. 2), compounds 1 and 3 emerged as the most effective inhibitors, suggesting that the hydroxy groups at C-5 and C-7, along with the methoxy group at C-4ʹ, play a crucial role in their activity. In contrast, compounds 2, 4, and 6 showed no inhibitory effects (IC50 > 100 μM), indicating that the presence of a hydroxy group at C-3ʹ, a methoxy group at C-6, or a glucose moiety at C-7 hindered inhibition. Additionally, compounds 7 and 9 demonstrated moderate inhibition, highlighting that the acetyl group at C-6ʹʹ significantly influences their inhibitory activity.
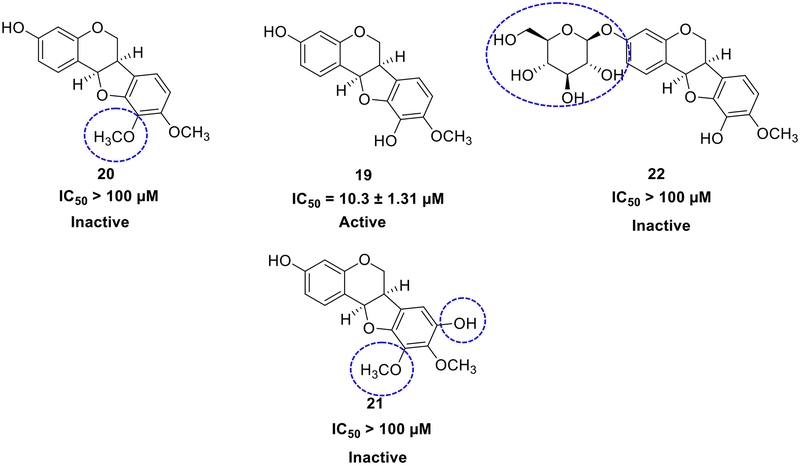
Furthermore, as showed in Fig. 3, among the pterocarpans, compound 19 emerged as the most potent inhibitor (IC50 10.3 ± 1.31 μM). In contrast, compounds 20, 21, 22, and 23 exhibit minimal to no effect on PTP1B. The potent inhibitory activity of 19 is attributed to its hydroxy groups at C-3 and C-10 and a methoxy group at C-9. The results also revealed that the methoxy group at C-10 (in 20, 21, and 23) and the glucose moiety at C-3 (in 22) diminished PTP1B inhibition. However, despite similar structural features, compound 24 exhibited moderate inhibitory activity (IC50 33.8 ± 5.61 μM), suggesting that the (E)-but-2-enoyl group at C-6ʹʹ enhances its ability to inhibit PTP1B.
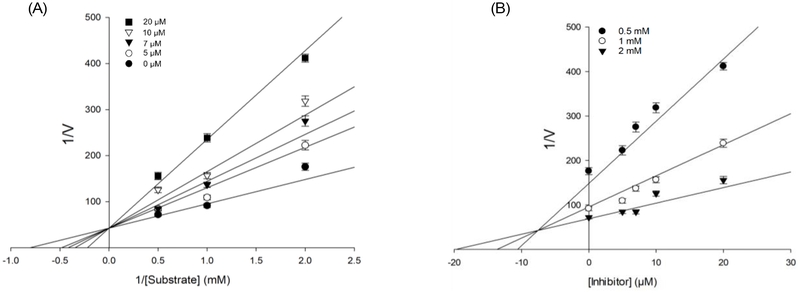
Building on the observed inhibitory effects and the existing data on the PTP1B inhibitory activity of compounds 1 and 3,22,23 this study conducted an enzyme kinetic assay to primarily elucidate the inhibition mode and inhibition constant (Ki) of compound 19. The type of inhibition was identified by analyzing the intersection points of trendlines, which illustrated the variations in inhibitory effects at different test sample concentrations across varying substrate levels. In particular, convergence at the x-axis indicated non-competitive inhibition, while intersection at the y-axis signified competitive mode. Alternatively, convergence within the x-y plane was indicative of mixed-type inhibition.43 As shown in Fig. 4A, the Lineweaver-Burk plot revealed that the trendlines converged at the y-axis, indicating that compound 19 inhibits PTP1B through a competitive mechanism. This finding suggested that compound 19 interacted with PTP1B at the active site, competing with the substrate at the same binding site, causing changes in the conformational dynamics of the enzymes, resulting in the diminishment of the enzyme’s function. Meanwhile, the Dixon plot (Fig. 4B) revealed the Ki value to be 7.6 ± 1.17 μM, represented by the intersection point of the lines on the x-axis. This Ki value corresponds to the dissociation constant of the inhibitor-enzyme complex, providing a measure of binding strength. A smaller Ki indicates a higher binding affinity, signifying that lower inhibitor concentrations are sufficient to suppress enzyme activity effectively.
Based on the enzyme kinetic analysis, molecular docking simulations were conducted to evaluate the binding interactions of compound 19 with PTP1B. The kinetic data demonstrated that compound 19 acts as a competitive inhibitor against PTP1B, suggesting that its docking site is localized within the protein's catalytic region (Fig. 5A).
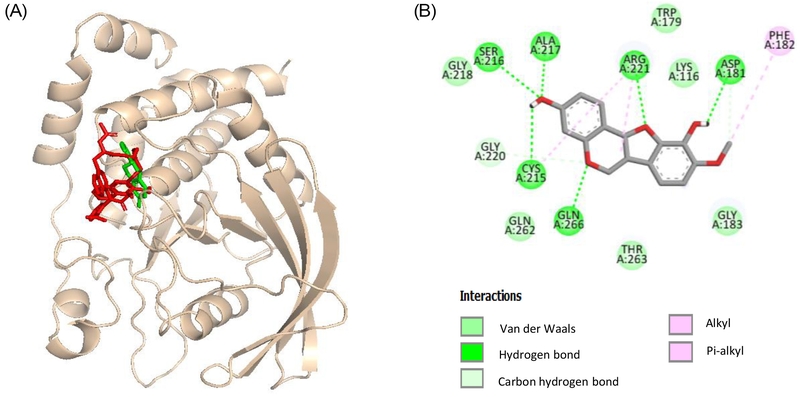
Molecular docking related PTP1B inhibition by compound C (red stick), 19 (green stick) (A), and 2D diagram of PTP1B inhibition by compound 19 (B).
As shown in Table 2, compound 19 demonstrated significant interactions with PTP1B, exhibiting a binding energy of −6.69 kcal/mol, surpassing compound C, a catalytic inhibitor, with a binding energy of −8.05 kcal/mol. These interactions were primarily mediated through van der Waals forces and hydrogen bonds. Specifically, van der Waals interactions were observed with residues Gly218, Gln262, Thr263, Gly183, Lys116, and Trp179. Hydrogen bonds were formed between the hydroxyl group at C-3 and residues Cys215, Ser216, and Ala217, as well as between the hydroxyl group at C-10 and Asp181. Additionally, the oxygen atoms in the cyclohexane and cyclopentane rings formed hydrogen bonds with Gln266 and Arg221, respectively. Furthermore, the interaction was strengthened by alkyl bonds with Phe182 and Arg221 and π-alkyl bonds with Arg221 and Cys215, further reinforcing the ligand-protein binding (Fig. 5B).

Binding energy and interactions of compound (19) and the reference inhibitor (compound C) with PTP1B
In summary, this study provides the first evidence of the therapeutic potential of flavonoids from the roots of A. membranaceus in diabetes treatment via PTP1B inhibition. Notably, formononetin (1), pratensein (3), and vesticarpan (19) exhibited significant inhibitory effects against PTP1B, underscoring the potential of the flavonoid fraction in diabetic therapy. In particular, identifying vesticarpan (19) as a novel PTP1B inhibitor serves as the foundation for further investigations to support the development of new therapeutic agents for diabetes treatment.
Acknowledgments
This research was supported by the National Research Foundation of Korea (NRF) funded by the Korea government (MSIT) (No. NRF-2020R1A5A2017323). The authors would like to thank Korea Basic Science Institute (KBSI) Daegu Center for the mass spectra measurement service.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
REFERENCES
-
Arroyave, F.; Montaño, D.; Lizcano, F. Int. J. Mol. Sci. 2020, 21, 8685.
[https://doi.org/10.3390/ijms21228685]

-
Krause, M.; De Vito, G. Nutrients 2023, 15, 4279.
[https://doi.org/10.3390/nu15194279]

- Zakir, M.; Ahuja, N.; Surksha, M. A.; Sachdev, R.; Kalariya, Y.; Nasir, M.; Kashif, M.; Shahzeen, F.; Tayyab, A.; Khan, M. S. M.; Junejo, M.; Kumar, F. M.; Varrassi, G.; Kumar, S.; Khatri, M.; Mohamad, T.; Cureus 2023, 15, e45835.
-
Rani, P. K.; Raman, R.; Gella, L.; Kulothungan, V.; Sharma, T. Middle East Afr. J. Ophthalmol. 2012, 19, 129–134.
[https://doi.org/10.4103/0974-9233.92129]

-
Bansal, D.; Gudala, K.; Muthyala, H.; Esam, H. P.; Nayakallu, R.; Bhansali, A. J. Diabetes Investig. 2014, 5, 714–721.
[https://doi.org/10.1111/jdi.12223]

-
Pathak, V.; Pathak, N. M.; O'Neill, C. L.; Guduric-Fuchs, J.; Medina, R. J. Clin. Med. Insights Endocrinol. Diabetes 2019, 12, 1179551419844521.
[https://doi.org/10.1177/1179551419844521]

-
Johnson, T. O.; Ermolieff, J.; Jirousek, M. R. Nat. Rev. Drug Discov. 2002, 1, 696–709.
[https://doi.org/10.1038/nrd895]

-
Cho, H. Vitam. Horm. 2013, 91, 405–424.
[https://doi.org/10.1016/B978-0-12-407766-9.00017-1]

-
Thiebaut, P. A.; Besnier, M.; Gomez, E.; Richard, V. J. Mol. Cell. Cardiol. 2016, 101, 50–57.
[https://doi.org/10.1016/j.yjmcc.2016.09.002]

-
Heinonen, K. M.; Tremblay, M. L. Cell Cycle 2006, 5, 1053–1056.
[https://doi.org/10.4161/cc.5.10.2735]

-
Liu, Z.; Gao, H.; Zhao, Z.; Huang, M.; Wang, S.; Zhan, J. Biomed. Pharmacother. 2023, 157, 113990.
[https://doi.org/10.1016/j.biopha.2022.113990]

-
Ullah, A.; Munir, S.; Badshah, S. L.; Khan, N.; Ghani, L.; Poulson, B. G.; Emwas, A.-H.; Jaremko, M. Molecules 2020, 25, 5243.
[https://doi.org/10.3390/molecules25225243]

-
AL-Ishaq, R. K.; Abotaleb, M.; Kubatka, P.; Kajo, K.; Büsselberg, D. Biomolecules 2019, 9, 430.
[https://doi.org/10.3390/biom9090430]

-
Yang, L.-P.; Shen, J.-G.; Xu, W.-C.; Li, J.; Jiang, J.-Q. Chem. Biodivers. 2013, 10, 1004–1054.
[https://doi.org/10.1002/cbdv.201100444]

-
Abdel-Latif, H. M. R.; Ahmed, H. A.; Shukry, M.; Chaklader, M. R.; Saleh, R. M.; Khallaf, M. A. Fishes 2022, 7, 319.
[https://doi.org/10.3390/fishes7060319]

-
Wang, H.-P.; Lin, Z.-Z.; Zhao, C.; Yin, Q.; Jia, J. J. Asian Nat. Prod. Res. 2024, 26, 1348–1357.
[https://doi.org/10.1080/10286020.2024.2366007]

-
Wu, Y.; Ou-Yang, J.-P.; Wu, K.; Wang, Y.; Zhou, Y.-F.; Wen, C.-Y. Acta Pharmacol. Sin. 2005, 26, 345–352.
[https://doi.org/10.1111/j.1745-7254.2005.00062.x]

-
Wang, N.; Zhang, D.; Mao, X.; Zou, F.; Jin, H.; Ouyang, J. Mol. Cell. Endocrinol. 2009, 307, 89–98.
[https://doi.org/10.1016/j.mce.2009.03.001]

-
Zhou, X.; Wang, L. L.; Tang, W. J.; Tang, B. J. Ethnopharmacol. 2021, 268, 113556.
[https://doi.org/10.1016/j.jep.2020.113556]

-
Wang, Z.; Li, X.-L.; Hong, K.-F.; Zhao, T.-T.; Dong, R.-X.; Wang, W.-M.; Li, Y.-T.; Zhang, G.-L.; Lin,-J.; Gui, D.-K.; Xu, Y.-H. Evid. Based Complement. Alternat. Med. 2021, 2021, 6675567.
[https://doi.org/10.1155/2021/6675567]

-
Lee, D.; Lee, D. H.; Choi, S.; Lee, J. S.; Jang, D. S.; Kang, K. S. Biomolecules 2019, 9, 618.
[https://doi.org/10.3390/biom9100618]

-
Wang, J.; Wang, L.; Han, L.; Han, Y.; Gu, J.; Chen, Z. Int. Immunopharmacol. 2024, 140, 112802.
[https://doi.org/10.1016/j.intimp.2024.112802]

-
Zhao, B. T.; Le, D. D.; Nguyen, P. H.; Ali, M. Y.; Choi, J.-S.; Min, B. S.; Shin, H. M.; Rhee, H. I.; Woo, M. H. Chem. Biol. Interact. 2016, 253, 27–37.
[https://doi.org/10.1016/j.cbi.2016.04.012]

-
Ha, M. T.; Lee, T. H.; Kim, C. S.; Prajapati, R.; Kim, J. A.; Choi, J. S.; Min, B. S. Phytochemistry 2022, 197, 113100.
[https://doi.org/10.1016/j.phytochem.2022.113100]

-
Li, S.-R.; Chen, P.-Y.; Chen, L.-Y.; Lo, Y.-F.; Tsai, I.-L.; Wang, E.-C. Tetrahedron Lett. 2009, 50, 2121–2123.
[https://doi.org/10.1016/j.tetlet.2009.02.159]

-
Salem, M. M.; Werbovetz, K. A. J. Nat. Prod. 2006, 69, 43–49.
[https://doi.org/10.1021/np0502600]

-
Lin, Y.-L.; Tsai, W.-J.; Chen, I.-S.; Kuo, Y. H. J. Chin. Chem. Soc. 1998, 45, 213–217.
[https://doi.org/10.1002/jccs.199800035]

-
Goto, H.; Terao, Y.; Akai, S. Chem. Pharm. Bull. 2009, 57, 346–360.
[https://doi.org/10.1248/cpb.57.346]

-
Velozo, L. S. M.; da Silva, B. P.; Bernardo, R. R.; Parente, J. P. Phytochemistry 1999, 52, 1473–1477.
[https://doi.org/10.1016/S0031-9422(99)00459-8]

-
Han, T.; Li, H.; Zhang, Q.; Zheng, H.; Qin, L. Chem. Nat. Compd. 2006, 42, 567–570.
[https://doi.org/10.1007/s10600-006-0215-2]

-
Barrero, A. F.; Sanchez, J. F.; Barrón, A.; Corrales, F.; Rodriguez, I. Phytochemistry 1989, 28, 161–164.
[https://doi.org/10.1016/0031-9422(89)85030-7]

-
Du, X.; Bai, Y.; Liang, H.; Wang, Z.; Zhao, Y.; Zhang, Q.; Huang, L. Magn. Reson. Chem. 2006, 44, 708–712.
[https://doi.org/10.1002/mrc.1806]

-
Zhang, L.-J.; Liu, H.-K.; Hsiao, P.-C.; Kuo, L.-M. Y.; Lee, I.-J.; Wu, T.-S.; Chiou, W.-F.; Kuo, Y.-H. J. Agric. Food Chem. 2011, 59, 1131–1137.
[https://doi.org/10.1021/jf103610j]

-
Pei, Y.; Li, R.; Fu, H.; Wang, J.; Zhou, Y. Fitoterapia 2007, 78, 602–604.
[https://doi.org/10.1016/j.fitote.2007.04.007]

-
Guo, K.; He, X.; Zhang, Y.; Li, X.; Yan, Z.; Pan, L.; Qin, B. Fitoterapia 2016, 114, 34–39.
[https://doi.org/10.1016/j.fitote.2016.08.009]

-
Ma, X.; Tu, P.; Chen, Y.; Zhang, T.; Wei, Y.; Ito, Y. J. Chromatogr. A. 2004, 1023, 311–315.
[https://doi.org/10.1016/j.chroma.2003.10.014]

-
Subarnas, A.; Oshima, Y.; Hikino, H. Phytochemistry 1991, 30, 2777–2780.
[https://doi.org/10.1016/0031-9422(91)85143-N]

- Radwan, M. M. Nat. Prod. Commun. 2008, 3, 1491–1494.
- Bojase, G.; Wanjala, C. C. W.; Majinda, R. R. T. Bull. Chem. Soc. Ethiop. 2001, 15, 131–136.
-
Ogata, T.; Yahara, S.; Hisatsune, R.; Konishi, R.; Nohara, T. Chem. Pharm. Bull. 1990, 38, 2750-2755.
[https://doi.org/10.1248/cpb.38.2750]

-
Piccinelli, A. L.; Campo Fernandez, M.; Cuesta-Rubio, O.; Márquez Hernández, I.; De Simone, F.; Rastrelli, L. J. Agric. Food. Chem. 2005, 53, 9010–9016.
[https://doi.org/10.1021/jf0518756]

-
Li, W.; Koike, K.; Asada, Y.; Hirotani, M.; Rui, H.; Yoshikawa, T.; Nikaido, T. Phytochemistry 2002, 60, 351–355.
[https://doi.org/10.1016/S0031-9422(02)00093-6]

-
Phong, N. V.; Oanh, V. T.; Yang, S. Y.; Choi, J. S.; Min, B. S.; Kim, J. A. Int. J. Biol. Macromol. 2021, 188, 719–728.
[https://doi.org/10.1016/j.ijbiomac.2021.08.091]