Phytochemical Investigation of Active Compounds from Celastrus orbiculatus Thunb. with α-Glucosidase Inhibitory Activity
Abstract
Diabetes, characterized by elevated blood glucose levels, has a significant impact on cardiovascular, neural, and vascular systems. α-Glucosidase inhibitors have emerged as potential therapeutic agents for type 2 diabetes, as they slow carbohydrate digestion and reduce postprandial blood sugar levels. In this study, we investigated the phytochemical and pharmacological properties of Celastrus orbiculatus Thunb., renowned for its diverse phytochemical constituents and potential medicinal applications. Through the application of chromatographic and spectroscopic techniques, we successfully isolated and structurally elucidated 16 compounds from the stems of C. orbiculatus. The in vitro α-glucosidase inhibitory activity of these compounds was evaluated. Notably, celaphanol A (1) and (+) lariciresinol (7) exhibited strong α-glucosidase inhibition, with IC50 values of 8.06 ± 0.30 and 48.02 ± 0.47 µM, respectively. Enzyme kinetics analysis revealed that the most active compound 1 acted as a non-competitive inhibitor against α-glucosidase, with a Ki value of 7.77 ± 0.16 µM. These findings underscore C. orbiculatus as a valuable source for discovering and developing new α-glucosidase inhibitors. Furthermore, compound 1 shows promise as a candidate for natural herbal therapy targeting α-glucosidase inhibition. This suggests the potential for further investigation into its effectiveness through in silico or in vivo studies using a diabetes model.
Keywords:
Celastrus orbiculatus, type 2 diabetes, α-glucosidase, enzyme kineticsIntroduction
Diabetes stands as a persistent metabolic disease marked by elevated levels of blood glucose.1,2 Over time, this condition inflicts notable damage upon the cardiac, neural, and vascular systems.3,4 The ailment encompasses two principal categories: type 1 diabetes arises from the incapacity of the pancreas to synthesize insulin,5 while type 2 diabetes emerges due to insufficient insulin production or inefficacious utilization of insulin by the body.6 This form accounts for more than 90% of diabetes mellitus instances.7,8
α-Glucosidase, a member of the glycoside hydrolase (GH) family, is usually manifest within the epithelium of the small intestine.9 Its function resides in the hydrolysis of disaccharides or oligosaccharides into monosaccharides, thus facilitating carbohydrate digestion.10 Inhibiting α-glucosidase has been examined as a therapeutic avenue for addressing type 2 diabetes, as this inhibition retards the liberation of sugar from starch and oligosaccharides.11,12 Consequently, it leads to a decelerated sugar absorption process and a reduction in postprandial blood sugar levels. Presently, α-glucosidase inhibitors such as acarbose, miglitol, and voglibose serve as efficacious clinical agents for blood sugar management and type 2 diabetes prevention.13 Nonetheless, these drugs entail gastrointestinal repercussions such as stomach discomfort and diarrhea.14 This highlights the need for discovering new α-glucosidase inhibitors that offer a lack of toxic side effects. This exploration for alternatives, especially from natural sources, has garnered substantial attention from researchers in recent times.
In recent decades, there has been a growing interest in exploring a wide range of natural products derived from plants due to their potential medicinal and therapeutic applications. Among these plants, Celastrus orbiculatus Thunb., commonly known as “oriental bittersweet” stands out for its rich phytochemical composition and interesting pharmacological properties.15 This climbing vine belongs to the Celastraceae family and is found distributed across various regions, with a history of traditional use in Asian folk medicine. The Great Dictionary of Chinese Medicine recorded that this plant was traditionally used to treat conditions such as rheumatoid arthritis, bruises, low back pain, injuries from falls, and amenorrhea.15,16 Similarly, in Korea, plant extracts are the main ingredients in remedies for rheumatoid arthritis, insomnia, and contusions.17 C. orbiculatus possesses a diverse range of phytochemicals, including alkaloids, terpenoids, flavonoids, lignans, and phenolic compounds. These compounds not only contribute to the plant ecological system but also hold the potential to exert various pharmacological effects on human health. Betulin-3β-yl-caffeate, a triterpene isolated from C. orbiculatus, has been reported to significantly inhibit osteoclast formation in a dose-dependent manner.18 At the molecular level, this compound inhibits the RANK-induced expression of c-Fos and the induction of nuclear factor of activated T cells 1, and downregulates mRNA expression of osteogenesis-associated marker genes, including tartrate-resistant acid phosphatase, dendritic cell-specific transmembrane protein, and matrix metalloprotein.18
Continuing our efforts to identify bioactive secondary metabolites from C. orbiculatus, 16 compounds were purified from the stems of this plant. The structures of all isolated compounds were elucidated using modern spectroscopic techniques. Furthermore, the in vitro α-glucosidase inhibitory activity of all isolated compounds from C. orbiculatus was investigated. Kinetic studies were conducted to clearly understand the mode of inhibition and the inhibition constant of active compound with the α-glucosidase enzyme.
Experimental
General experimental procedures – 1H- and 13C-NMR data were obtained on a Bruker Avance Digital 500 MHz spectrometer (Bruker, Karlsruhe, Germany) using tetramethylsilane as internal standard, J in Hz. Compound isolation was conducted on column chromatography (CC) using silica gel 60 (0.063-0.200 mm), LiChroprep RP-18 (40-63 µm), and MCI gel CHP20P (75-150 µm) (Merck, Darmstadt, Germany). High performance liquid chromatography (HPLC) was conducted on Waters Alliance HPLC system (MA, USA) equipped with a 1525 Binary pump, Water 2998 PDA, and YMC Pack ODS column (20 × 250 mm, 4 µm). Thin-layer chromatography was performed using glass plates pre-coated with silica gel 60F254 and RP-18 F254s (Merck). Spots were detected under UV light at 254 and 356 nm, and visualized by spraying the plates with 10% H2SO4 followed by heating at 110oC for 1-2 min. α-Glucosidase from Saccharomyces cerevisiae (EC 3.2.1.20, G5003), acarbose (A8980), and 4-nitrophenyl α-D-glucopyranoside (N1377) were supplied from Sigma-Aldrich (St. Louis, MO, USA).
Plant material – Dried stems of C. orbiculatus were collected in the Daegu Catholic University herbal garden in January 2019. Plant authentication was confirmed by one of the authors, Professor Byung Sun Min. The voucher specimen (accession code: 21B-CO) was kept at the Pharmacognosy Lab., College of Pharmacy, Kyungpook National University.
Extraction and isolation – The dried stems of C. orbiculatus (8.0 kg) were extracted three times using MeOH (16 L for 4 hours each) under reflux conditions. The resulting extract was concentrated under in vacuo. The MeOH residue (400 g) was mixed with 1.2 L of distilled water and sequentially partitioned with n-hexane and EtOAc.
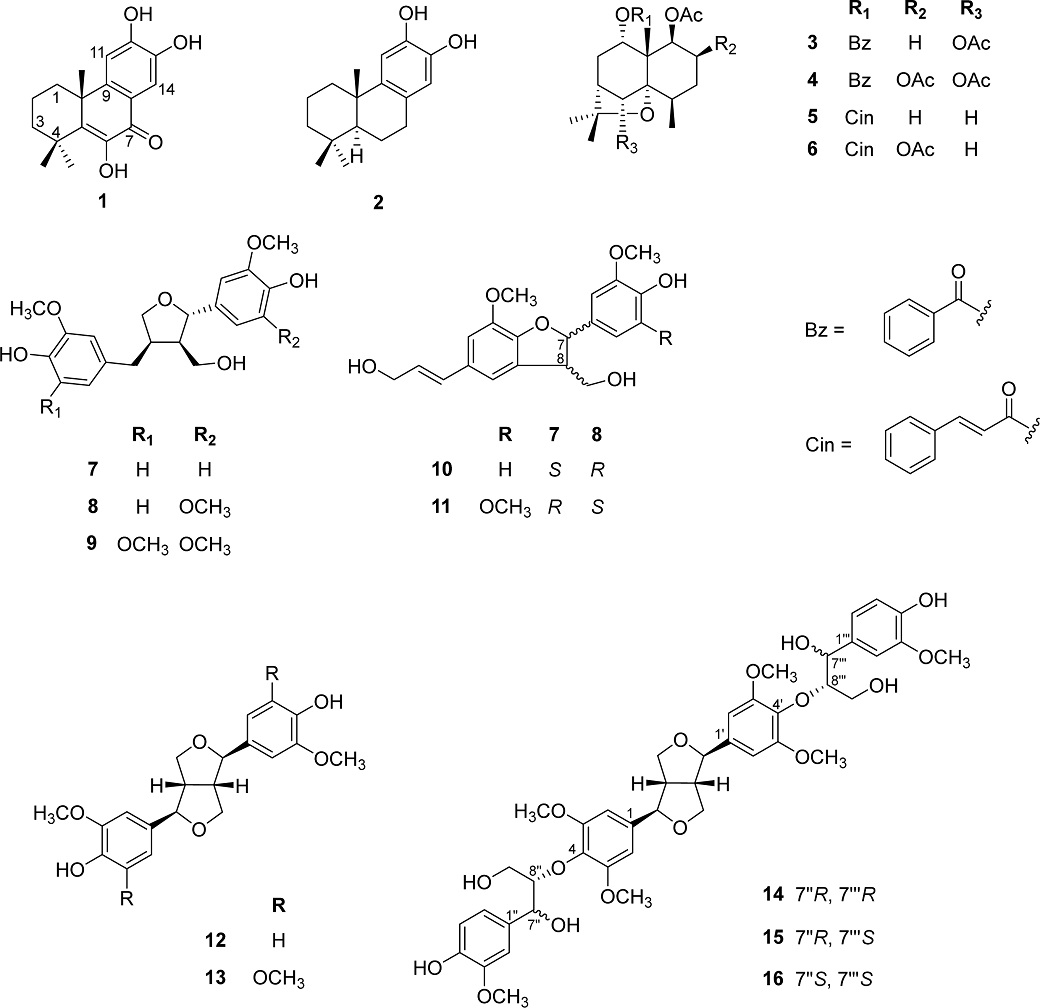
The n-hexane extract (78.3 g) was subjected to silica gel vacuum liquid chromatography (VLC) eluted with n-hexane:acetone (gradient 10:1 → 0:1, v/v) to obtain six fractions (1A–1F). Fraction 1D (9.2 g) was separated by silica gel CC eluted with CH2Cl2:MeOH (gradient 50:1 → 0:1, v/v) to obtain 11 fractions (2A–2K). Fraction 2E (1.3 g) was purified by RP-18 CC eluted with MeOH:H2O (9:1, v/v) to isolate compounds 1 (69.7 mg) and 2 (6.5 mg). From fraction 2F (446.6 mg), compounds 3 (1.8 mg), 4 (10.2 mg), 5 (2.0 mg), and 6 (5.5 mg) were isolated by RP-18 CC eluting with MeOH:H2O (gradient 19:1 → 9:1, v/v).
The EtOAc extract (40.1 g) was chromatographed on silica gel VLC eluted with CH2Cl2:MeOH (gradient 99:1 → 0:1, v/v) to obtain 13 fractions (3A–3M). From fraction 3E (1.9 g), compounds 7 (2.0 mg), 8 (2.0 mg), and 9 (2.0 mg) were isolated by MCI CC eluting with MeOH:H2O (1:1, v/v). Fraction 3F (2.0 g) was chromatographed on MCI CC eluting with MeOH:H2O (1:1, v/v), followed by silica gel CC, using CH2Cl2:acetone (4:1, v/v) as mobile phase, to yield compounds 10 (10.5 mg), 11 (5.8 mg), 12 (6.2 mg), and 13 (5.5 mg). Compounds 14 (2.3 mg), 15 (2.0 mg), and 16 (2.1 mg) were isolated from fraction 3J (327.5 mg) by silica gel CC, using CH2Cl2:MeOH (25:1, v/v) as eluent, followed by preparative HPLC with an isocratic mixture solvent 50% ACN in H2O (6 mL/min, 60 min).
Celaphanol A (1) – Amorphous powder; 1H-NMR (CDCl3, 500 MHz): δH 7.11 (1H, s, H-11), 7.71 (1H, s, H-14), 1.38 (3H, s, H-15), 1.39 (3H, s, H-16), 1.42 (3H, s, H-17); 13C-NMR (CDCl3, 125 MHz): δC 33.4 (C-1), 17.5 (C-2), 37.7 (C-3), 36.2 (C-4), 142.9 (C-5), 143.7 (C-6), 180.2 (C-7), 120.3 (C-8), 150.7 (C-9), 40.9 (C-10), 112.1 (C-11), 150.9 (C-12), 144.2 (C-13), 111.9 (C-14), 27.5 (C-15), 28.1 (C-16), 34.8 (C-17).19
(+)-7-Deoxynimbidiol (2) – White amorphous powder; 1H-NMR (CDCl3, 500 MHz): δH 6.76 (1H, s, H-11), 6.52 (1H, s, H-14), 1.15 (3H, s, H-15), 0.93 (3H, s, H-16), 0.91 (3H, s, H-17); 13C-NMR (CDCl3, 125 MHz): δC 39.3 (C-1), 19.5 (C-2), 41.8 (C-3), 33.5 (C-4), 50.9 (C-5), 19.2 (C-6), 30.0 (C-7), 128.1 (C-8), 141.2 (C-9), 37.5 (C-10), 111.6 (C-11), 143.5 (C-12), 141.5 (C-13), 115.3 (C-14), 33.4 (C-15), 25.0 (C-16), 21.7 (C-17).20
1α,6β-Diacetoxy-9β-benzoyloxydihydro-β-agarofuran (3) – Amorphous powder; 1H-NMR (CDCl3, 500 MHz): δH 1.01 (3H, d, J = 7.3 Hz, H-14), 1.33 (3H, s, H-15), 1.40 (3H, s, H-12), 1.41 (3H, s, H-13), 1.45 (1H, m, H-3b), 1.59 (1H, m, H-2b), 1.88 (1H, m, H-2a), 2.18 (1H, m, H-8b), 2.20 (1H, m, H-3a), 2.21 (1H, m, H-7), 2.26 (1H, m, H-4), 2.42 (1H, ddd, J = 16.2, 7.1, 3.1 Hz, H-8a), 5.01 (1H, d, J = 6.9 Hz, H-9), 5.32 (1H, s, H-6), 5.46 (1H, dd, J = 12.0, 4.3 Hz, H-1), OAc [1.61, 2.11 (each 3H, s)], OBz [7.44 (2H, t, J = 7.8 Hz), 7.55 (1H, t, J = 7.8 Hz), 8.07 (2H, dd, J = 8.2, 1.1 Hz)]; 13C-NMR (CDCl3, 125 MHz): δC 73.5 (C-1), 21.7 (C-2), 26.9 (C-3), 34.1 (C-4), 90.0 (C-5), 79.7 (C-6), 49.0 (C-7), 32.2 (C-8), 73.5 (C-9), 50.7 (C-10), 82.7 (C-11), 26.2 (C-12), 30.8 (C-13), 17.6 (C-14), 19.0 (C-15), OAc (21.0, 21.6, 170.1, 170.4), OBz (128.4, 128.5, 130.2, 133.4, 165.7).21
Triptogelin C1 (4) – Amorphous powder; 1H-NMR (CDCl3, 500 MHz): δH 1.22 (3H, d, J = 7.6 Hz, H-14), 1.41 (3H, s, H-12), 1.41 (3H, s, H-13), 1.48 (3H, s, H-15), 1.81 (1H, m, H-3b), 2.22 (1H, m, H-8b), 2.38 (1H, m, H-4), 2.42 (1H, m, H-3a), 2.47 (1H, m, H-8a), 2.48 (1H, m, H-7), 4.96 (1H, d, J = 6.9 Hz, H-9), 5.42 (1H, s, H-6), 5.58 (1H, s, H-2), 5.59 (1H, s, H-1), OAc [1.61, 2.03, 2.13 (each 3H, s)], OBz [7.43 (2H, dd, J = 10.7, 4.8 Hz), 7.56 (1H, t, J = 7.8 Hz), 8.04 (2H, dd, J = 8.3, 1.2 Hz)]; 13C-NMR (CDCl3, 125 MHz): δC 71.2 (C-1), 70.0 (C-2), 31.0 (C-3), 34.2 (C-4), 89.6 (C-5), 79.2 (C-6), 48.9 (C-7), 31.6 (C-8), 73.1 (C-9), 50.0 (C-10), 82.9 (C-11), 26.9 (C-12), 30.7 (C-13), 18.6 (C-14), 20.4 (C-15), OAc (20.5, 21.5, 21.6, 169.7, 170.2, 170.3), OBz (128.4, 129.5, 130.2, 133.4, 165.6).22
Celafolins B-1 (5) – Amorphous powder; 1H-NMR (CDCl3, 500 MHz): δH 1.26 (3H, d, J = 7.4 Hz, H-14), 1.20 (3H, s, H-12), 1.20 (3H, s, H-13), 1.38 (3H, s, H-15), 4.82 (1H, d, J = 6.4 Hz, H-9), 5.43 (1H, dd, J = 12.0, 4.3 Hz, H-1), OAc [1.83 (3H, s)], OCin [6.42 (1H, d, J = 16.0 Hz), 7.68 (1H, d, J = 16.0 Hz), 7.35–7.56 (5H, m)]; 13C-NMR (CDCl3, 125 MHz): δC 73.6 (C-1), 22.0 (C-2), 27.0 (C-3), 40.4 (C-4), 87.4 (C-5), 36.5 (C-6), 43.9 (C-7), 31.6 (C-8), 73.8 (C-9), 49.7 (C-10), 82.5 (C-11), 26.3 (C-12), 30.4 (C-13), 17.6 (C-14), 18.8 (C-15), OAc (21.0, 170.1), OCin (117.9, 128.5, 128.8, 130.5, 134.5, 145.3, 166.5).23
1α,2α,8β-Triacetoxy-9β-cinnamoyloxy-β-dihidro agarofuran (6) – Amorphous powder; 1H-NMR (CDCl3, 500 MHz): δH 1.24 (3H, d, J = 8.0 Hz, H-14), 1.22 (3H, s, H-12), 1.41 (3H, s, H-13), 1.54 (3H, s, H-15), 5.07 (1H, d, J = 6.1 Hz, H-9), 5.39 (1H, dd, J = 6.1, 2.9 Hz, H-8), 5.52 (1H, s, H-1), 5.52 (1H, s, H-2), OAc [1.82, 1.94, 2.03 (each 3H, s)], OCin [6.42 (1H, d, J = 16.0 Hz), 7.66 (1H, d, J = 16.0 Hz), 7.34–7.56 (5H, m)]; 13C-NMR (CDCl3, 125 MHz): δC 70.5 (C-1), 70.8 (C-2), 31.2 (C-3), 39.3 (C-4), 86.9 (C-5), 35.9 (C-6), 48.6 (C-7), 70.5 (C-8), 72.3 (C-9), 47.6 (C-10), 82.5 (C-11), 24.9 (C-12), 31.2 (C-13), 19.3 (C-14), 19.9 (C-15), OAc (20.5, 20.7, 21.1, 170.0, 170.1, 170.1), OCin (117.9, 128.4, 128.9, 130.3, 134.4, 145.3, 166.4).24
(+) Lariciresinol (7) – Amorphous powder; 1H-NMR (CD3OD, 500 MHz): δH 2.40 (1H, m, H-8), 2.52 (1H, dd, J = 13.4, 11.4 Hz, H-7′a), 2.76 (1H, m, H-8′), 2.96 (1H, dd, J = 13.4, 4.8 Hz, H-7′b), 3.66 (1H, dd, J = 10.9, 6.5 Hz, H-9a), 3.75 (1H, dd, J = 8.4, 5.9 Hz, H-9′a), 3.86 (1H, dd, J = 10.9, 8.0 Hz, H-9b), 4.01 (1H, dd, J = 8.4, 6.4 Hz, H-9′b), 4.74 (1H, d, J = 6.9 Hz, H-7), 6.67 (1H, dd, J = 8.0, 1.9 Hz, H-6′), 6.74 (1H, d, J = 8.0 Hz, H-5′), 6.79 (1H, m, H-6), 6.79 (1H, m, H-5), 6.82 (1H, d, J = 1.9 Hz, H-2′), 6.93 (1H, d, J = 1.4 Hz, H-2), OCH3 [3.87, 3.85 (each 3H, s)]; 13C-NMR (CD3OD, 125 MHz): δC 135.7 (C-1), 110.7 (C-2), 149.0 (C-3), 147.2 (C-4), 116.0 (C-5), 119.8 (C-6), 84.1 (C-7), 54.1 (C-8), 60.5 (C-9), 133.5 (C-1′), 113.4 (C-2′), 149.0 (C-3′), 145.9 (C-4′), 116.2 (C-5′), 122.2 (C-6′), 33.7 (C-7′), 43.9 (C-8′), 73.5 (C-9′), 56.4 (2×OCH3).25
5′-Methoxylariciresinol (8) – Amorphous powder; 1H-NMR (CD3OD, 500 MHz): δH 2.41 (1H, m, H-8), 2.55 (1H, dd, J = 13.4, 11.4 Hz, H-7′a), 2.74 (1H, m, H-8′), 2.91 (1H, dd, J = 13.1, 5.3 Hz, H-7′b), 3.76 (1H, dd, J = 8.7, 5.7 Hz, H-9′a), 3.79 (1H, dd, J = 11.0, 6.9 Hz, H-9a), 3.93 (1H, dd, J = 10.9, 7.0 Hz, H-9b), 4.06 (1H, dd, J = 8.6, 6.4 Hz, H-9′b), 4.80 (1H, d, J = 6.2 Hz, H-7), 6.57 (2H, s, H-2, 6), 6.68 (2H, m, H-2′, 6′), 6.84 (1H, d, J = 8.5 Hz, H-5′), 3×OCH3 [3.85 (each 3H, s)]; 13C-NMR (CD3OD, 125 MHz): δC 133.5 (C-1), 104.3 (C-2, 6), 149.3 (C-3, 5), 133.5 (C-4), 84.3 (C-7), 54.1 (C-8), 60.5 (C-9), 133.5 (C-1′), 113.4 (C-2′), 146.5 (C-3′), 144.0 (C-4′), 114.4 (C-5′), 122.1 (C-6′), 33.7 (C-7′), 43.8 (C-8′), 73.6 (C-9′), 56.8, 56.4 (3×OCH3).26
(7S, 8R, 8′R)-5,5′-Dimethoxylariciresinol (9) – Amorphous powder; 1H-NMR (CD3OD, 500 MHz): δH 2.41 (1H, m, H-8), 2.53 (1H, dd, J = 13.4, 11.3 Hz, H-7′a), 2.77 (1H, m, H-8′), 2.97 (1H, dd, J = 13.5, 4.9 Hz, H-7′b), 3.78 (1H, dd, J = 8.5, 6.1 Hz, H-9′a), 3.69 (1H, dd, J = 11.0, 6.6 Hz, H-9a), 3.90 (1H, m, H-9b), 4.03 (1H, dd, J = 8.6, 6.5 Hz, H-9′b), 4.79 (1H, d, J = 6.7 Hz, H-7), 6.53 (2H, s, H-2, 6), 6.65 (2H, s, H-2′, 6′), 4×OCH3 [3.85, 3.86 (each 3H, s)]; 13C-NMR (CD3OD, 125 MHz): δC 132.8 (C-1), 104.3 (C-2, 6), 149.3 (C-3, 5), 135.0 (C-4), 84.2 (C-7), 54.2 (C-8), 60.5 (C-9), 134.9 (C-1′), 107.0 (C-2′, 6′), 149.3 (C-3′, 5′), 136.0 (C-4′), 34.2 (C-7′), 43.8 (C-8′), 73.6 (C-9′), 56.8 (4×OCH3).27
(+)-Dehydrodiconiferyl alcohol (10) – Amorphous powder; 1H-NMR (CD3OD, 500 MHz): δH 3.52 (1H, dd, J = 12.3, 6.2 Hz, H-8), 3.81 (1H, dd, J = 10.9, 6.9 Hz, H-9a), 3.85 (1H, m, H-9b), 4.23 (2H, dd, J = 6.0, 1.6 Hz, H-9′), 5.55 (1H, d, J = 6.3 Hz, H-7), 6.26 (1H, dt, J = 15.8, 5.9 Hz, H-8′), 6.57 (1H, d, J = 15.8 Hz, H-7′), 6.80 (1H, d, J = 8.1 Hz, H-5), 6.86 (1H, dd, J = 8.2, 2.0 Hz, H-6), 6.98 (2H, m, H-2, 6′), 7.00 (1H, d, J = 2.0 Hz, H-2′), 2×OCH3 [3.84, 3.90 (each 3H, s)]; 13C-NMR (CD3OD, 125 MHz): δC 134.6 (C-1), 110.6 (C-2), 149.1 (C-3), 147.6 (C-4), 116.2 (C-5), 119.8 (C-6), 89.3 (C-7), 55.2 (C-8), 64.9 (C-9), 130.4 (C-1′), 112.1 (C-2′), 145.5 (C-3′), 149.3 (C-4′), 132.6 (C-5′), 116.5 (C-6′), 132.0 (C-7′), 127.6 (C-8′), 63.9 (C-9′), 56.8, 56.4 (3×OCH3).28
(−)-Simulanol (11) – Amorphous powder; 1H-NMR (CD3OD, 500 MHz): δH 3.40 (1H, q, J = 6.3 Hz, H-8), 3.68 (1H, dd, J = 10.9, 6.5 Hz, H-9a), 3.75 (1H, m, H-9b), 4.22 (2H, dd, J = 6.0, 1.4 Hz, H-9′), 5.55 (1H, d, J = 6.3 Hz, H-7), 6.25 (1H, dt, J = 15.8, 5.9 Hz, H-8′), 6.56 (1H, d, J = 15.8 Hz, H-7′), 6.70 (2H, s, H-2, 6), 6.97 (1H, s, H-6′), 6.98 (1H, s, H-2′), 3×OCH3 [3.83, 3.91 (each 3H, s)]; 13C-NMR (CD3OD, 125 MHz): δC 133.8 (C-1), 104.2 (C-2, 6), 149.4 (C-3, 5), 133.8 (C-4), 89.4 (C-7), 55.3 (C-8), 64.9 (C-9), 132.7 (C-1′), 112.2 (C-2′), 145.5 (C-3′), 149.2 (C-4′), 130.3 (C-5′), 116.5 (C-6′), 132.0 (C-7′), 127.6 (C-8′), 63.9 (C-9′), 56.8 (3×OCH3).29
Pinoresinol (12) – Amorphous powder; 1H-NMR (CD3OD, 500 MHz): δH 6.93 (2H, d, J = 1.7 Hz, H-2, 2′), 6.75 (2H, d, J = 8.0 Hz, H-5, 5′), 6.79 (2H, dd, J = 8.0, 1.7 Hz, H-6, 6′), 4.69 (2H, d, J = 4.4 Hz, H-7, 7′), 3.12 (2H, m, H-8, 8′), 3.83 (2H, dd, J = 8.9, 3.6 Hz, H-9b, 9′b), 4.21 (2H, dd, J = 8.9, 6.8 Hz, H-9a, 9′a), 2×OCH3 [3.84 (each 3H, s)]; 13C-NMR (CD3OD, 125 MHz): δC 133.8 (C-1, 1′), 111.0 (C-2, 2′), 149.1 (C-3, 3′), 147.3 (C-4, 4′), 116.1 (C-5, 5′), 120.1 (C-6, 6′), 87.5 (C-7, 7′), 55.4 (C-8, 8′), 72.6 (C-9, 9′), 56.4 (2×OCH3).30
Syringaresinol (13) – Amorphous powder; 1H-NMR (CD3OD, 500 MHz): δH 6.67 (4H, s, H-2, 2′, 6, 6′), 4.80 (2H, d, J = 4.0 Hz, H-7, 7′), 3.14 (2H, m, H-8, 8′), 3.64 (2H, dd, J = 9.0, 3.6 Hz, H-9b, 9′b), 4.34 (2H, dd, J = 9.0, 6.9 Hz, H-9a, 9′a), 4×OCH3 [3.86 (each 3H, s)]; 13C-NMR (CD3OD, 125 MHz): δC 132.2 (C-1, 1′), 103.7 (C-2, 2′, 6, 6′), 148.4 (C-3, 3′, 5, 5′), 135.4 (C-4, 4′), 86.6 (C-7, 7′), 54.5 (C-8, 8′), 71.7 (C-9, 9′), 55.9 (4×OCH3).31
Hedyotisol A (14) – Amorphous powder; 1H-NMR (CD3OD, 500 MHz): δH 6.72 (4H, s, H-2, 2′, 6, 6′), 4.78 (2H, d, J = 3.3 Hz, H-7, 7′), 3.12 (2H, m, H-8, 8′), 3.91 (2H, m, H-9b, 9′b), 4.28 (2H, m, H-9a, 9′a), 7.00 (2H, d, J = 1.7 Hz, H-2″, 2‴), 6.75 (2H, d, J = 8.2 Hz, H-5″, 5‴), 6.87 (2H, dd, J = 8.2, 1.7 Hz, H-6″, 6‴), 4.99 (2H, s, H-7″, 7‴), 4.16 (2H, m, H-8″, 8‴), 3.45 (2H, m, H-9″b, 9‴b), 3.85 (2H, m, H-9″a, 9‴a), 6×OCH3 [3.89, 3.85, 3.84 (each 3H, s)]; 13C-NMR (CD3OD, 125 MHz): δC 138.9 (C-1, 1′), 104.2 (C-2, 2′, 6, 6′), 154.3 (C-3, 3′, 5, 5′), 135.6 (C-4, 4′), 87.3 (C-7, 7′), 55.7 (C-8, 8′), 74.1 (C-9, 9′), 135.8 (C-1″, 1‴), 111.5 (C-2″, 2‴), 148.7 (C-3″, 3‴), 147.2 (C-4″, 4‴), 115.7 (C-5″, 5‴), 120.8 (C-6″, 6‴), 73.1 (C-7″, 7‴), 88.7 (C-8″, 8‴), 61.7 (C-9″, 9‴), 56.7, 56.3 (6×OCH3).32
Hedyotisol B (15) – Amorphous powder; 1H-NMR (CD3OD, 500 MHz): δH 6.72 (4H, s, H-2, 2′, 6, 6′), 4.78 (2H, d, J = 3.3 Hz, H-7, 7′), 3.12 (2H, m, H-8, 8′), 3.91 (2H, m, H-9b, 9′b), 4.28 (2H, m, H-9a, 9′a), 7.00 (2H, d, J = 1.7 Hz, H-2″, 2‴), 6.75 (2H, d, J = 8.2 Hz, H-5″, 5‴), 6.87 (2H, dd, J = 8.2, 1.7 Hz, H-6″, 6‴), 4.99 (2H, d, J = 6.9 Hz, H-7″, 7‴), 4.16 (1H, m, H-8″), 3.45 (1H, m, H-9″b), 3.85 (1H, m, H-9″), 3.96 (1H, m, H-8‴), 3.33 (1H, m, H-9‴b), 3.65 (1H, m, H-9‴a), 6×OCH3 [3.89, 3.85, 3.84 (each 3H, s)]; 13C-NMR (CD3OD, 125 MHz): δC 138.9 (C-1, 1′), 104.2 (C-2, 2′, 6, 6′), 154.3 (C-3, 3′, 5, 5′), 135.6 (C-4, 4′), 87.3 (C-7, 7′), 55.7 (C-8, 8′), 74.1 (C-9, 9′), 133.8 (C-1″), 111.5 (C-2″), 148.7 (C-3″), 147.2 (C-4″), 115.7 (C-5″), 120.8 (C-6″), 73.1 (C-7″), 87.3 (C-8″), 61.7 (C-9″), 133.5 (C-1‴), 111.6 (C-2‴), 148.7 (C-3‴), 146.9 (C-4‴), 115.8 (C-5‴), 120.8 (C-6‴), 73.1 (C-7‴), 88.7 (C-8‴), 61.8 (C-9‴) 56.7, 56.3 (6×OCH3).32
Hedyotisol C (16) – Amorphous powder; 1H-NMR (CD3OD, 500 MHz): δH 6.72 (4H, s, H-2, 2′, 6, 6′), 4.78 (2H, d, J = 3.3 Hz, H-7, 7′), 3.12 (2H, m, H-8, 8′), 3.91 (2H, m, H-9b, 9′b), 4.28 (2H, m, H-9a, 9′a), 7.00 (2H, d, J = 1.7 Hz, H-2″, 2‴), 6.78 (2H, d, J = 8.2 Hz, H-5″, 5‴), 6.87 (2H, dd, J = 8.2, 1.7 Hz, H-6″, 6‴), 4.91 (2H, dd, J = 5.4, 2.7 Hz, H-7″, 7‴), 3.96 (2H, m, H-8″, 8‴), 3.33 (2H, m, H-9″b, 9‴b), 3.63 (2H, m, H-9″a, 9‴a), 6×OCH3 [3.86, 3.84 (each 3H, s)]; 13C-NMR (CD3OD, 125 MHz): δC 138.9 (C-1, 1′), 104.3 (C-2, 2′, 6, 6′), 154.6 (C-3, 3′, 5, 5′), 136.2 (C-4, 4′), 87.3 (C-7, 7′), 55.6 (C-8, 8′), 74.1 (C-9, 9′), 136.2 (C-1″, 1‴), 111.4 (C-2″, 2‴), 148.8 (C-3″, 3‴), 147.3 (C-4″, 4‴), 115.8 (C-5″, 5‴), 120.7 (C-6″, 6‴), 73.1 (C-7″, 7‴), 87.3 (C-8″, 8‴), 61.7 (C-9″, 9‴), 56.7, 56.3 (6×OCH3).32
Inhibitory activity on α-glucosidase – The in vitro assay for α-glucosidase inhibition was conducted in accordance with previous protocols.33,34 The enzymatic reaction was initiated by introducing 130 μL of α-glucosidase enzyme (at a concentration of 0.16 unit/mL) into a phosphate buffer with a pH of 6.8, containing 100 μM substrate. This reaction mixture was placed within a 96-well plate and supplemented with 20 μL of MeOH, or test compounds were dissolved in MeOH. Subsequently, the mixture was incubated at 37oC along with 50 μL of 4-nitrophenyl α-D-glucopyranoside (at a concentration of 1 mM).
The production of 4-nitrophenol resulting from the hydrolysis of 4-nitrophenyl α-D-glucopyranoside was quantified by measuring absorbance at 405 nm using a microplate spectrophotometer (BioTek Epoch 2, Agilent, CA, USA). Acarbose, an α-glucosidase inhibitor, was used as a positive control. α-Glucosidase inhibitory activity (%) was determined by applying the following equation:
Inhibitory activity (%) = [(ΔA−ΔB) / ΔA] × 100, where ΔA and ΔB represent the signal intensities of the control and inhibitor solutions after a 20 min incubation period, respectively.
The IC50 values were subjected to analysis and calculation using SigmaPlot 10.0 (Systat Software, CA, USA).
Enzyme kinetics with α-glucosidase – The investigation of the inhibition mode of α-glucosidase by active compounds involved the application of two complementary kinetic techniques: Lineweaver–Burk and Dixon plots.35 The experimental results were subjected to analysis and visualization using SigmaPlot 10.0. Dixon plot, also known as single reciprocal plot, was employed to evaluate enzymatic reactions across various concentrations of test compounds. This was achieved by observing the effects of various substrate concentrations (0.625, 1.25, and 2.5 mM). To attain a more comprehensive insight, a dual reciprocal Lineweaver–Burk plot was generated. Within this plot, the α-glucosidase inhibition mode was identified at different substrate concentrations, both in the presence and absence of different concentrations of the test compound (0, 2.95, 8.3, and 19.5 µM). The inhibition constant (Ki) was calculated using the Dixon plot, with the value of the x-axis or [I] axis taken as -Ki.
Statistical analysis – The results were represented as the means ± standard error of the mean (SEM) for three independent experiments. Statistical significance (p < 0.05) was calculated using Duncan′s tests and ANOVA.
Results and Discussion
Stems of C. orbiculatus were extracted with MeOH to obtain methanol residue. Following solvent removal under reduced pressure, the resulting methanol residue was suspended in water and subjected to partitioning using n-hexane and EtOAc. The n-hexane and EtOAc extracts were separated by CC and preparative HPLC, leading to the isolation of 16 compounds, including two diterpenes (1–2), four sesquiterpenes (3–6), and ten lignans (7–16) (Fig. 1). Through comprehensive analysis of spectroscopic data, including 1H and 13C-NMR, as well as comparison with those reported in the literature, the chemical structures of all isolated compounds were successfully elucidated as celaphanol A (1),19 (+)-7-deoxynimbidiol (2),20 1α,6β-diacetoxy-9β-benzoyloxydihydro-β-agarofuran (3),21 triptogelin C1 (4),22 celafolins B-1 (5),23 1α,2α,8β-triacetoxy-9β-cinnamoyloxy-β-dihidro agarofuran (6),24 (+) lariciresinol (7),25 5′-methoxylariciresinol (8),26 (7S, 8R, 8′R)-5,5′-dimethoxylariciresinol (9),27 (+)-dehydrodiconiferyl alcohol (10),28 (−)-simulanol (11),29 pinoresinol (12),30 syringaresinol (13),31 and hedyotisols A–C (14–16).32 Compounds 7–11 were isolated from C. orbiculatus for the first time, whereas compounds 14–16 had previously been obtained in the Celastraceae family.36 This study also reported the first isolation of compounds 14–16 from this plant.
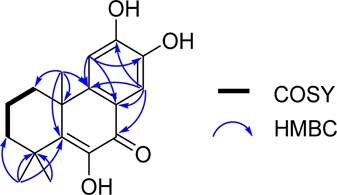
Compound 1 was obtained as an amorphous powder. The 1H-NMR data of 1 showed signals of three methyl groups at δH 1.38 (3H, s, H-15), 1.39 (3H, s, H-16), and 1.42 (3H, s, H-17), and two aromatic protons at δH 7.11 (1H, s, H-11) and 7.71 (1H, s, H-14). They correlated to the carbons resonating at δC 27.5, 28.1, 34.8, 112.1, and 111.9, respectively. In addition, the 13C-NMR, DEPT and HMQC spectra also revealed signals of a carbonyl group at δC 180.2 (C-7), a double bond at δC 142.9 (C-5) and 143.7 (C-6), three methylene groups at δC 33.4 (C-1), 17.5 (C-2), and 37.7 (C-3), and two quaternary carbons at δC 36.2 (C-4) and 40.9 (C-10). The position of carbonyl group was determined by HMBC correlations of δH 7.71 (H-14) with δC 180.2 (C-7), 120.3 (C-8), 150.7 (C-9), and 150.9 (C-12) (Fig. 2). The above analysis indicates signals characteristic of a diterpene skeleton, and through a comparison with data found in the literature, the structure of compound 1 was determined to be celaphanol A.19
All isolated compounds (1–16) were examined for their ability to inhibit α-glucosidase activity. As a comparison, acarbose was used as a positive control and exhibited an IC50 value of 215.69 ± 1.55 µM. Among isolated diterpenes, compound 1 exhibited the most potent inhibitory activity on α-glucosidase, with an IC50 value of 8.06 ± 0.30 µM (Table 1). The absence of a hydroxy group at C-6 and a carbonyl group at C-7 in compound 2, in contrast to compound 1, resulted in no inhibitory effect against α-glucosidase, highlighting the significance of the functional groups in α-glucosidase inhibition. Compound 7 exhibited a strong inhibitory effect on α-glucosidase, with an IC50 value of 48.02 ± 0.47 µM. Compounds 8 and 9, with one and two additional methoxy groups compared to compound 7, showed no α-glucosidase inhibition. This evidence suggests that the α-glucosidase inhibitory activity could be affected by the number of less polar substituents, such as -OCH3 groups, present in the chemical structure of enterolignans. Unfortunately, the remaining compounds exhibited weak or no α-glucosidase inhibitory effects, with IC50 values > 100 µM.
Enzyme kinetics investigations were conducted to understand the mode of inhibition and determine the inhibition constant (Ki value), which correlates with the interactions between the active compound and the target enzyme. This understanding was obtained using graphical methods, particularly the Lineweaver–Burk and Dixon techniques.35 The Lineweaver–Burk plot revealed that the point of intersection of the inhibitor lines on either the x-axis or y-axis corresponded to non-competitive or competitive inhibition, respectively.
As shown in Fig. 3, compound 1 displayed a series of intersecting straight lines on the x-axis or 1/V-axis. This observation indicates that this active compound functions as a non-competitive α-glucosidase inhibitor. The Ki value for compound 1 was calculated to be 7.77 ± 0.16 µM. A lower Ki value signifies a lower concentration required for the formation of the enzyme-inhibitor complex.37-39 Consequently, compounds with lower Ki values have consistently demonstrated higher effectiveness as α-glucosidase inhibitors.33,40
In summary, a total of 16 secondary metabolites (1–16) were successfully isolated from the dried stems of C. orbiculatus. The chemical structures of these isolated compounds were elucidated using advanced spectroscopy techniques, including 1H- and 13C-NMR, and were further confirmed through comparison with published literature. Among these compounds, celaphanol A (1) displayed the most potent inhibitory effect on α-glucosidase, exhibiting an IC50 value of 8.06 ± 0.30 µM. Through enzyme kinetics analysis, it was revealed that the active compound 1 functioned as a non-competitive inhibitor against α-glucosidase, with a Ki value of 7.77 ± 0.16 µM. This study contributes not only to the growing diversity of secondary metabolites but also offers valuable insights into the α-glucosidase inhibitory activity of compounds from C. orbiculatus. Furthermore, our findings strongly indicate the potential of active compound 1 as a natural α-glucosidase inhibitory therapy.
Acknowledgments
This research was supported by the National Research Foundation of Korea (NRF) funded by the Korea government (MSIT) (No. NRF-2020R1A5A2017323). The authors would like to thank Korea Basic Science Institute (KBSI) Daegu Center for the mass spectra measurement service.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
-
Viroonluecha, P.; Egea-Lopez, E.; Santa, J. PLos One 2022, 17, e0274608.
[https://doi.org/10.1371/journal.pone.0274608]

-
Le, T. T.; Ha, M. T.; Hoang, L. M.; Vu, N. K.; Kim, J. A.; Min, B. S. Nat. Prod. Sci. 2022, 28, 143-152.
[https://doi.org/10.20307/nps.2022.28.3.143]

-
Saw, E. L.; Pearson, J. T.; Schwenke, D. O.; Munasinghe, P. E.; Tsuchimochi, H.; Rawal, S.; Coffey, S.; Davis, P.; Bunton, R.; Van Hout, I.; Kai, Y.; Williams, M. J. A.; Kakinuma, Y.; Fronius, M.; Katare, R. Cardiovasc. Diabetol. 2021, 20, 50.
[https://doi.org/10.1186/s12933-021-01231-8]

-
Feener, E. P.; King, G. L. Lancet 1997, 350, S9-S13.
[https://doi.org/10.1016/S0140-6736(97)90022-2]

-
DiMeglio, L. A.; Evans-Molina, C.; Oram, R. A. Lancet 2018, 391, 2449-2462.
[https://doi.org/10.1016/S0140-6736(18)31320-5]

-
Berbudi, A.; Rahmadika, N.; Tjahjadi, A. I.; Ruslami, R. Curr. Diabetes Rev. 2020, 16, 442-449.
[https://doi.org/10.2174/1573399815666191024085838]

-
Phong, N. V.; Oanh, V. T.; Yang, S. Y.; Choi, J. S.; Min, B. S.; Kim, J. A. Int. J. Biol. Macromol. 2021, 188, 719-728.
[https://doi.org/10.1016/j.ijbiomac.2021.08.091]

- Hariftyani, A. S.; Kurniawati, L. A.; Khaerunnisa, S.; Veterini, A. S.; Setiawati, Y.; Awaluddin, R. Nat. Prod. Sci. 2021, 27, 99-114.
-
Hamid, H. A.; Yusoff, M. M.; Liu, M.; Karim, M. R. J. Funct. Foods 2015, 16, 74-80.
[https://doi.org/10.1016/j.jff.2015.04.011]

-
Ćorković, I.; Gašo-Sokač, D.; Pichler, A.; Šimunović, J.; Kopjar, M. Life 2022, 12, 1692.
[https://doi.org/10.3390/life12111692]

-
Dirir, A. M.; Daou, M.; Yousef, A. F.; Yousef, L. F. Phytochem. Rev. 2022, 21, 1049-1079.
[https://doi.org/10.1007/s11101-021-09773-1]

-
Striegel, L.; Kang, B.; Pilkenton, S. J.; Rychlik, M.; Apostolidis, E. Front. Nutr. 2015, 2, 3.
[https://doi.org/10.3389/fnut.2015.00003]

-
van de Laar, F. A.; Lucassen, P. L.; Akkermans, R. P.; van de Lisdonk, E. H.; Rutten, G. E.; van Weel, C. Diabetes Care 2005, 28, 154-163.
[https://doi.org/10.2337/diacare.28.1.154]

-
Min, S. H.; Yoon, J. H.; Hahn, S.; Cho, Y. M. J. Diabetes Investig. 2018, 9, 893-902.
[https://doi.org/10.1111/jdi.12754]

-
Shen, Y.; Chen, B.-L.; Zhang, Q.-X.; Zheng, Y.-Z.; Fu, Q. J. Ethnopharmacol. 2019, 241, 111934.
[https://doi.org/10.1016/j.jep.2019.111934]

-
Feng, X.; Yin, Z.; Ou, S.; Chu, Z.; Feng, J.; Luo, Y.; Hu, Y.; Liu, Y.; Jiang, W.; Wang, X.; Wang, H. J. Ethnopharmacol. 2023, 310, 116363.
[https://doi.org/10.1016/j.jep.2023.116363]

-
Jeon, H. Nat. Prod. Sci. 2011, 17, 135-141.
[https://doi.org/10.1038/nm0211-141b]

-
Vu, T. O.; Tran, P. T.; Seo, W.; Lee, J. H.; Min, B. S.; Kim, J. A. J. Nat. Med. 2021, 75, 56-65.
[https://doi.org/10.1007/s11418-020-01444-3]

-
Chen, B.; Duan, H.; Takaishi, Y. Phytochemistry 1999, 51, 683-687.
[https://doi.org/10.1016/S0031-9422(99)00079-5]

-
Xiong, Y.; Wang, K.; Pan, Y.; Sun, H.; Tu, J. Bioorg. Med. Chem. Lett. 2006, 16, 786-789.
[https://doi.org/10.1016/j.bmcl.2005.11.023]

-
Takaishi, Y.; Ohshima, S.; Nakano, K.; Tomimatsu, T.; Tokuda, H.; Nishino, H.; Iwashima, A. J. Nat. Prod. 1993, 56, 815-824.
[https://doi.org/10.1021/np50096a003]

-
Takaishi, Y.; Tokura, K.; Tamai, S.; Ujita, K.; Nakano, K.; Tomimatsu, T. Phytochemistry 1991, 30, 1567-1572.
[https://doi.org/10.1016/0031-9422(91)84210-J]

-
Tu, Y. Q.; Wang, D. Z.; Zhang, H. J.; Lin, Z. Phytochemistry 1991, 30, 271-273.
[https://doi.org/10.1016/0031-9422(91)84136-G]

- Wang, M. A.; Chen, F. H. Chem. J. Chinese Universities 1996, 17, 1250-1252.
-
Xie, L.-H.; Akao, T.; Hamasaki, K.; Deyama, T.; Hattori, M. Chem. Pharm. Bull (Tokyo). 2003, 51, 508-515.
[https://doi.org/10.1248/cpb.51.508]

-
Duh, C. Y.; Phoebe, C. H. Jr.; Pezzuto, J. M.; Kinghorn, A. D.; Farnsworth, N. R. J. Nat. Prod. 1986, 49, 706-709.
[https://doi.org/10.1021/np50046a031]

-
Choi, S. U.; Yang, M. C.; Lee, K. H.; Kim, K. H.; Lee, K. R. Arch. Pharm. Res. 2007, 30, 1067-1074.
[https://doi.org/10.1007/BF02980239]

-
Yeo, H.; Chin, Y.-W.; Park, S.-Y.; Kim, J. Arch. Pharm. Res. 2004, 27, 287-290.
[https://doi.org/10.1007/BF02980061]

-
Yang, Y.-P.; Cheng, M.-J.; Teng, C.-M.; Chang, Y.-L.; Tsai, I.-L.; Chen, I.-S. Phytochemistry 2002, 61, 567-572.
[https://doi.org/10.1016/S0031-9422(02)00268-6]

-
Moon, S.-S.; Rahman, A. A.; Kim, J.-Y.; Kee, S.-H. Bioorg. Med. Chem. 2008, 16, 7264-7269.
[https://doi.org/10.1016/j.bmc.2008.06.032]

-
Cai, X. F.; Lee, I. S.; Dat, N. T.; Shen, G.; Kang, J. S.; Kim, D. H.; Kim, Y. H. Arch. Pharm. Res. 2004, 27, 738-741.
[https://doi.org/10.1007/BF02980142]

-
Xiong, L.; Zhu, C.; Li, Y.; Tian, Y.; Lin, S.; Yuan, S.; Hu, J.; Hou, Q.; Chen, N.; Yang, Y.; Shi, J. J. Nat. Prod. 2011, 74, 1188-1200.
[https://doi.org/10.1021/np200117y]

-
Phong, N. V.; Yang, S. Y.; Min, B. S.; Kim, J. A. J. Mol. Struct. 2023, 1282, 135188.
[https://doi.org/10.1016/j.molstruc.2023.135188]

-
Phong, N. V.; Gao, D.; Kim, J. A.; Yang, S. Y. Metabolites 2023, 13, 557.
[https://doi.org/10.3390/metabo13040557]

-
Coleman, M. H. Nature 1965, 205, 798-799.
[https://doi.org/10.1038/205798a0]

-
Zhu, J. X.; Ren, J.; Qin, J. J.; Cheng, X. R.; Zeng, Q.; Zhang, F.; Yan, S. K.; Jin, H. Z.; Zhang, W. D. Arch. Pharm. Res. 2012, 35, 1739-1747.
[https://doi.org/10.1007/s12272-012-1005-y]

-
Phong, N. V.; Min, B. S.; Yang, S. Y.; Kim, J. A. Appl. Sci. 2022, 12, 10685.
[https://doi.org/10.3390/app122010685]

-
Holdgate, G. A.; Meek, T. D.; Grimley, R. L. Nat. Rev. Drug Discov. 2018, 17, 115-132.
[https://doi.org/10.1038/nrd.2017.219]

-
Tonge, P. J. ACS Infect. Dis. 2019, 5, 796-808.
[https://doi.org/10.1021/acsinfecdis.9b00012]

-
Daou, M.; Elnaker, N. A.; Ochsenkühn, M. A.; Amin, S. A.; Yousef, A. F.; Yousef, L. F. PLos One 2022, 17, e0264969.
[https://doi.org/10.1371/journal.pone.0264969]