A New Bicyclic Megastigmane Derivative from By-product of Ginseng Berry Extract
Abstract
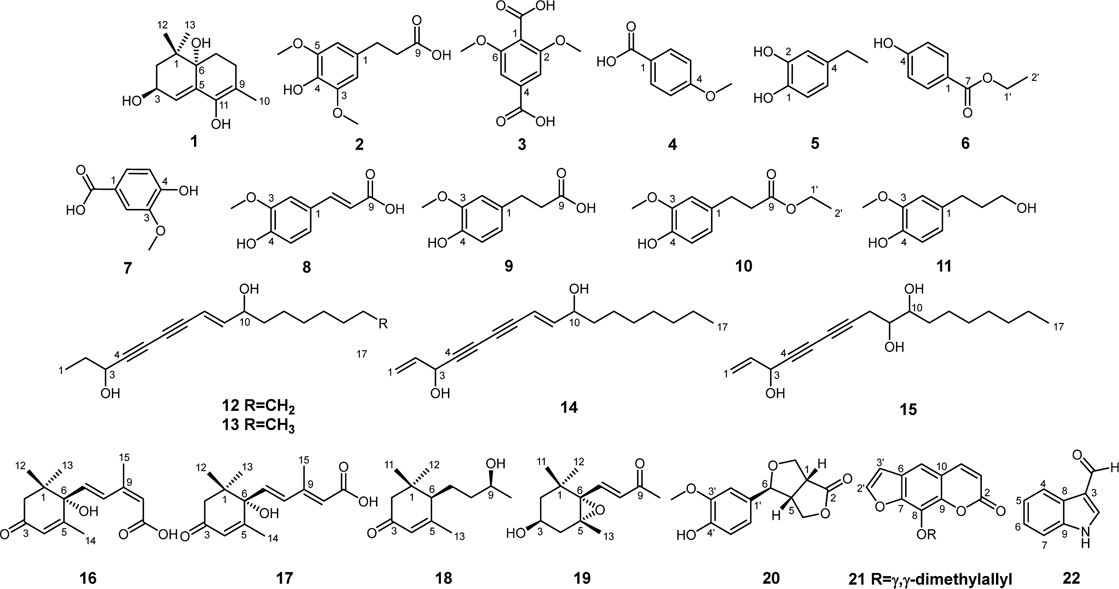
A new bicyclic megastigmane derivative, (3S,6S)-amoreol (1), was isolated and determined along with twenty known compounds, dihydrosinapic acid (2), 2,6-dimethoxyterephthalic acid (3), 4-methoxybenzoic acid (4), 4-ethylbenzene-1,2-diol (5), ethylparaben (6), vanillic acid (7), ferulic acid (8), 3-(4-hydroxy-3-methoxyphenyl)propionic acid (9), ethyl dihydroferulate (10), dihydroconiferyl alcohol (11), 1,2-dihydrodendroarboreol B (12), 1,2-dihydropanaxydiol (13), panaxydiol (14), panaxytriol (15), (S)-cis-abscisic acid (16), (R)-trans-abscisic acid (17), (6R,9S)-blumenol C (18), (3S,5R,6S,7E)-5,6-epoxy-3-hydroxy-7-megastigmene-9-one (19), (+)-salicifoliol (20), imperatorin (21), indole-3-carboxaldehyde (22) from the by-product of ginseng berry extract. The structures of the isolated compounds were determined through spectroscopic techniques including UV, 1D-, and 2D NMR, CD, MS and optical rotation. The absolute configuration of 1 was elucidated based on the experimental and calculated electronic circular dichroism (ECD) spectra.
Keywords:
Panax ginseng, Araliaceae, Megastigmane, (3S,6S)-amoreolIntroduction
Panax ginseng C.A. Meyer, a representative medicinal plant in the East belonging to the Araliaceae family, has been cultivated for thousands of years in East Asian countries such as Korea, China, and Japan. The genus name “Panax” originates from the Greek words “Pan” (all) and “Axos” (healing), meaning “healing of all” “panacea”.1-3
Panax ginseng has been reported to possess various pharmacological effects, including immune enhancement, anticancer properties, anti-aging benefits, potent antioxidant activity, and blood pressure regulation. These effects are mainly attributed to its active compounds known as ginsenosides, which are triterpene glycosides.4 To date, around 6,000 reports on ginsenosides have been documented,5 with approximately 50 types of ginsenosides identified.4 In addition to ginsenosides, P. ginseng contains various compounds, including polysaccharides, polyacetylenes, flavonoids, phenolic compounds, and essential oils.6 Non-saponin compounds have also shown pharmacological effects, among which panaxytriol, a unique polyacetylene compound found in ginseng, displays antitumor properties.7,8 Furthermore, Ginseng polysaccharides have been reported to promote intestinal health, regulate metabolism, and improve the balance of gut microbiota.9,10
As the economic and industrial value of P. ginseng continues to rise, its cultivation is experiencing a notable increase in South Korea. In a study conducted by Fan et al. (2023), the annual yield of P. ginseng exceeds 20,000 tons.11 However, as the ginseng industry grows, the disposal of ginseng by-products after the production of commercial ginseng products has become a major concern in recent years, and many efforts are being made to find a good way to recycle or upcycle these by-products.
Until now, P. ginseng research has primarily focused on the root, while the fruits and leaves have been considered less valued.12 However, it has been discovered that ginseng berry contains higher levels of ginsenoside Re compared to those of the root,13 leading to increase in studies exploring the phytochemical composition and physiological effects of ginseng berry.14-16 Moreover, many companies are leveraging ginseng berry to launch diverse commercial products, and there are efforts to reutilize the by-products after processing ginseng berry. To determine the value of the by-products from ginseng berry, scientific evidence such as chemical analysis and biological evaluation should be carried out.
In this study, we focused on the phytochemical study of the industrial by-products of ginseng berry extract, and identified 22 compounds, including 5 phenylpropanoids (2, 8-11), 5 simple phenolic compounds (3-7), 4 polyacetylenes (12-15), 2 sesquiterpenes (16-17), 3 nor-sesquiterpenoids (1 and 18-19), 1 furolactone-type lignan (20), 1 coumarin (21), and 1 indole derivative (22). The chemical analysis of isolates was performed through spectroscopic evidence such as UV, 1D-, and 2D NMR, CD, MS and optical rotation.
Experimental
General experimental procedures – The NMR spectra were obtained using an AVANCE 500 spectrometer (Bruker, Karlsruhe, Germany) at 500 MHz for 1H NMR, and 125 MHz for 13C NMR. The HPLC analysis was carried out with a 1260 Infinity HPLC system (Agilent Technologies, Santa Clara, CA, USA). The isolation of compounds was performed using an HPLC system from Gilson (Gilson, WI, USA), equipped with binary pumps, a UV detector, and a liquid handler. Optical rotation and Circular Dichromism spectra were recorded with a P-2000 polarimeter (Jasco, Tokyo, Japan) and J-815 spectrometer (Jasco, Tokyo, Japan). The UV-1800 Shimadzu UV spectrophotometer was used for measuring UV spectra. Column chromatography, employing Silica gel 60 (40-63 μm, Merck, Germany), ZEOprep 90 C18 (40-63 μm, Zeochme, Uetikon, Switzerland), and Diaion HP-20 (Mistubishi chemical, Tokyo, Japan) as the stationary phases, was employed for the separation and purification of compounds. An Agilent 6530 Q-TOF LC/MS spectrometer (Agilent Technologies, Santa Clara, CA, USA) was used to record the mass spectra. The HPLC analysis was carried out with a Phenomenex Luna® C18 column (4.6 × 250 mm ID., 5 µm; Phenomenex, Torrance, CA, USA), and the HPLC separation was performed using a Luna® C18(2) column (21.2 × 250 mm I.D., 5 mm; Phenomenex, USA).
The organic solvents required for chromatography were obtained from Dae-Jung Chemicals & Metals Co. Ltd. (Gyunggi-do, Korea), and HPLC grade solvents, such as acetonitrile and methanol, were purchased from JT Baker Scientific Korea (Seoul, Korea). Deionized water was obtained by utilizing the Millipore Milli-Q® water purification system.
Sample materials – The by-product of the ginseng berry extract (non-saponin extract) was provided by Amorepacific Co. The manufacturing process of the by-product was not included in this paper due to Amorepacific’s internal regulations.
Extraction and isolation – The by-product of the ginseng berry extract (300 g) was suspended in 90% MeOH and then partitioned with n-hexane. The solvent of each layer was removed under reduced pressure to yield n-hexane soluble- and 90% MeOH soluble extracts. The 90% MeOH (174 g) soluble extract was subjected to Diaion HP-20 column chromatography (C.C.) using 70% MeOH (3 L) and obtained 17 g of dry residue. The 70% MeOH fraction was subjected to a silica gel C.C. [n-hexane-EtOAc (2:1, 1:1, v/v), and MeOH] to yield 13 subfractions (E1-E13). E12 (1.4 g) was separated using reversed phase (RP) C.C. with MeOH-H2O (30:70, v/v) to give 14 subfractions (E12.1-E12.14). E12.10 (33.1 mg) was purified by RP-HPLC using MeCN-H2O (30:70, v/v) to give compounds 20 (1.7 mg) and 3 (5 mg). Compound 2 (17.1 mg) was isolated from E12.12 (73.7 mg) by RP-HPLC eluted with MeCN-H2O (20:80, v/v). E7 (500 mg) was subjected to NP-MPLC silica gel C.C. with n-hexane-EtOAc (30:1, 4:1, 3:1, 2:1, v/v) to afford 16 subfractions (E7.1-E7.16). E7.13 (18.4 mg) was purified by RP-HPLC using MeCN-H2O (30:70→100:0, v/v) to yield compounds 7 (1.5 mg) and 15 (18.2 mg). Compound 21 (0.4 mg) was obtained by RP-HPLC with MeCN-H2O (75:25, v/v) from subfraction E7.15 (22 mg). E10 (1.1 g) was partitioned over ODS column eluted with MeOH-H2O (55:45, v/v) to yield 16 sub-fractions (E10.16-E10.16). Compounds 17 (2.5 mg), 19 (4.6 mg), and 22 (1.6 mg) were purified from E10.6 (38.3 mg) by RP-HPLC using MeCN-H2O (38:62, v/v). E10.4 (96 mg) was purified by preparative HPLC and eluted with MeCN-H2O (25:75, v/v) to yield compounds 8 (26.5 mg), 9 (27.6 mg), and 11 (22.2 mg). E10.7 (61.5 mg) was isolated by preparative HPLC and eluted by MeCN-H2O (45:55, v/v), giving compounds 16 (5.8 mg) and 1 (3.1 mg). Compound 18 (1.9 mg) was obtained from subfraction E10.9 (49.7 mg) by RP-HPLC using MeCN-H2O (40:60, v/v). E1.2 (843.7 mg) was chromatographed over silica gel C.C eluted with n-hexane-EtOAc (5:1, v/v) to afford seven subfractions (E1.2.1-E1.2.7). E1.2.2 (421.8 mg) was fractionated by reverse-phase column chromatography with MeCN and H2O (35:65→40:60, v/v) to obtain nine subfractions (E1.2.2.1-E1.2.2.9). E1.2.2.5 (21.0 mg) was subjected to RP-HPLC using MeOH-H2O (60:40, v/v) as mobile phase to furnish compounds 4 (5.0 mg) and 5 (6.0 mg). Compound 6 (0.3 mg) was obtained from E1.2.2.6 (10.3 mg) by preparative HPLC utilizing MeCN-H2O (30:70, v/v). Purification of compound 10 (13.3 mg) was carried out by RP-HPLC of E1.2.2.7 (35.9 mg) with mobile phase of MeCN-H2O (60:40, v/v). E1.2.4 (70.7 mg) was purified by preparative HPLC eluted with MeCN-H2O (80:20, v/v) to yield compounds 12 (1.6 mg), 13 (1.1 mg), and 14 (13.4 mg).
(3S,6S)-Amoreol (1) − White powder; C13H20O3; [a]25D = +20.4° (c 0.12, MeOH); CD (MeOH): Δε (nm) = -1.21 (245), +1.91 (205); UV (MeOH) λmax (log ε): 208.6 (4.16); ESI-Q-TOF-MS: m/z 223.1332 [M-H]-; 1H-NMR (CDCl3, 500 MHz): δ 6.88 (1H, dd, J = 2.7, 1.7 Hz, H-4), 4.43 (1H, ddd, J = 9.8, 6.4, 2.7 Hz, H-3), 2.44 – 2.24 (2H, m, H-7a, 8a), 2.04 (1H, m, H-8b), 1.94 (1H, ddd, J = 13.3, 6.4, 1.7 Hz, H-2a), 1.87 (1H, ddd, J = 12.7, 9.3, 5.0 Hz, H-7b), 1.65 (3H, s, H-10), 1.51 (1H, dd, J = 13.3, 9.8 Hz, H-2b), 1.06 (3H, s, H-13), 1.03 (3H, s, H-12); 13C-NMR (CDCl3, 125 MHz): δ 163.48 (C-11), 137.63 (C-4), 132.40 (C-5), 110.82 (C-9), 87.70 (C-6), 65.72 (C-3), 44.99 (C-2), 38.36 (C-8), 37.44 (C-1), 31.20 (C-7), 24.24 (C-12), 23.63 (C-10), 22.59 (C-13).
Dihydrosinapic acid (2) − Yellow crystal; C11H12O5; ESI-Q-TOF-MS: m/z 249.0737 [M+Na]+; 1H-NMR (CDCl3, 500 MHz): δ 6.41 (2H, s, H-2, 6), 3.84 (6H, s, 3-OCH3, 5-OCH3), 2.87 (2H, t, J = 7.7 Hz, H-7), 2.64 (2H, t, J = 7.7 Hz, H-8); 13C-NMR (CDCl3, 125 MHz): δ 179.04 (C-9), 147.22 (C-3, 5), 133.41 (C-4), 131.46 (C-1), 105.13 (C-2, 6), 56.46 (3-OCH3, 5-OCH3), 36.19 (C-8), 31.05 (C-7).
Syringic acid (3) − Yellow crystal; C10H10O6; ESI-Q-TOF-MS: m/z 197.0447 [M-H]-; 1H-NMR (CD3OD, 500 MHz): δ 7.33 (2H, s, H-3, 5), 3.88 (6H, s, 2-OCH3, 6-OCH3); 13C-NMR (CD3OD, 125 MHz): δ 170.10 (4-COOH), 148.97 (C-2, 6), 141.88 (C-4), 122.07 (C-1), 108.47 (C-3, 5), 56.91 (2-OCH3, 6-OCH3).
4-Methoxybenzoic acid (4) − White crystalline solid; C8H8O3; ESI-Q-TOF-MS: m/z 151.0393 [M-H]-; 1H-NMR (CD3OD, 500 MHz): δ 7.87 (2H, m, H-2, 6), 6.82 (2H, m, H-3, 5), 3.84 (3H, s, 4-OCH3); 13C-NMR (CD3OD, 500 MHz): δ 168.86 (1-COOH), 163.66 (C-4), 132.88 (C-2, 6), 122.37 (C-1), 116.29 (C-3, 5), 52.37 (4-OCH3).
4-Ethylbenzene-1,2-diol (5) − Light brown oil; C8H10O2; ESI-Q-TOF-MS: m/z 137.0606 [M-H]-; 1H-NMR (CD3OD, 500 MHz): δ 6.65 (1H, d, J = 8.0 Hz, H-6), 6.62 (1H, d, J = 2.1 Hz, H-3), 6.49 (1H, dd, J = 8.0, 2.1 Hz, H-5), 2.47 (2H, q, J = 7.6 Hz, H-5), 1.16 (3H, t, J = 7.6 Hz, H-6); 13C-NMR (CD3OD, 125 MHz): δ 146.20 (C-2), 144.18 (C-1), 137.40 (C-4), 120.08 (C-5), 116.38 (C-3), 116.08 (C-6), 29.34 (C-5), 16.60 (C-6).
Ethylparaben (6) − White solid; C9H10O3; ESI-Q-TOF-MS: m/z 167.0704 [M+H]+; 1H-NMR (CD3OD, 500 MHz): δ 7.87 (1H, m, H-2, 6), 6.82 (1H, m, H-3, 5), 4.31 (1H, q, J = 7.1 Hz, H-1'), 1.36 (2H, t, J = 7.1 Hz, H-2'); 13C-NMR (CD3OD, 125 MHz): δ 168.40 (C-7), 163.63 (C-4), 132.83 (C-2, 6), 122.64 (C-1), 116.25 (C-3, 5), 61.82 (C-1'), 14.81 (C-2').
Vanillic acid (7) − White-yellowish powder; C8H8O4; ESI-Q-TOF-MS: m/z 169.0499 [M+H]+; 1H-NMR (CDCl3, 500 MHz): δ 7.70 (1H, dd, J = 8.3, 1.9 Hz, H-6), 7.57 (1H, d, J = 1.9 Hz, H-2), 6.95 (1H, d, J = 8.3 Hz, H-5), 3.95 (3H, s, 3-OCH3); 13C-NMR (CDCl3, 125 MHz): δ 170.23 (1-COOH), 150.96 (C-4), 145.91 (C-3), 125.38 (C-6), 121.30 (C-1), 114.40 (C-5), 112.27 (C-2), 56.34 (3-OCH3).
Ferulic acid (8) − White amorphous powder; C10H10O4; ESI-Q-TOF-MS: m/z 217.0475 [M+Na]+; 1H-NMR (CD3OD, 500 MHz): δ 7.60 (1H, d, J = 15.9 Hz, H-7), 7.18 (1H, d, J = 2.0 Hz, H-2), 7.06 (1H, dd, J = 8.2, 2.0 Hz, H-6), 6.81 (1H, d, J = 8.2 Hz, H-5), 7.60 (1H, d, J = 15.9 Hz, H-8), 3.89 (3H, s, 3-OCH3); 13C-NMR (CD3OD, 125 MHz): δ 171.12 (C-9), 150.64 (C-4), 149.50 (C-3), 147.08 (C-7), 127.91 (C-1), 124.14 (C-6), 116.58 (C-5), 116.04 (C-8), 111.78(C-2), 55.0 (3-OCH3).
3-(4-Hydroxy-3-methoxyphenyl)propionic acid (9) − Colorless solid; C10H12O4; ESI-Q-TOF-MS: m/z 197.0813 [M+H]+; 1H-NMR (CD3OD, 500 MHz): δ 6.79 (1H, d, J = 2.0 Hz, H-2), 6.70 (1H, d, J = 8.0 Hz, H-5), 6.64 (1H, dd, J = 8.0, 2.0 Hz, H-6), 3.82 (3H, s, 3-OCH3), 2.82 (2H, t, J = 7.7 Hz, H-7), 2.55 (2H, dd, J = 8.1, 7.2 Hz, H-8); 13C-NMR (CD3OD, 125 MHz): δ 177.10 (C-9), 149.00 (C-3), 145.97 (C-4), 133.84 (C-1), 121.78 (C-6), 116.26 (C-5), 113.12 (C-2), 56.43 (3-OCH3), 37.32 (C-8), 31.83 (C-7).
Ethyl dihydroferulate (10) − Pale brown oil; C12H16O4; ESI-Q-TOF-MS: m/z 225.1118 [M+H]+; 1H-NMR (CD3OD, 500 MHz): δ 6.77 (1H, d, J = 2.0 Hz, H-2), 6.69 (1H, d, J = 8.0 Hz, H-5), 6.62 (1H, dd, J = 8.0, 2.0 Hz, H-6), 4.09 (2H, q, J = 7.1 Hz, H-1'), 3.82 (3H, s, 3-OCH3), 2.82 (2H, t, J = 7.6 Hz, H-5), 2.57 (2H, t, J = 7.6 Hz, H-8), 1.21 (3H, t, J = 7.1 Hz, H-2'); 13C-NMR (CD3OD, 125 MHz): δ 175.06 (C-9), 149.03 (C-3), 146.08 (C-4), 133.62 (C-1), 121.86 (C-6), 116.29 (C-5), 113.21 (C-2), 61.62 (C-1'), 56.49 (3-OCH3), 37.46 (C-8), 31.81 (C-7), 14.65 (C-2').
Dihydroconiferyl alcohol (11) − Colorless crystals; C10H14O3; ESI-Q-TOF-MS: m/z 205.0845 [M+Na]+; 1H-NMR (CD3OD, 500 MHz): δ 6.76 (1H, d, J = 2.0 Hz, H-2), 6.69 (1H, d, J = 8.0 Hz, H-5), 6.62 (1H, dd, J = 8.0, 2.0 Hz, H-6), 3.82 (3H, s, 3-OCH3), 3.55 (2H, t, J = 6.5 Hz, H-9), 2.58 (2H, dd, J = 8.6, 6.9 Hz, H-7), 1.79 (2H, m, H-8); 13C-NMR (CD3OD, 125 MHz): δ 148.94 (C-3), 145.60 (C-4), 135.06 (C-1), 121.89 (C-6), 116.17 (C-5), 113.22 (C-2), 62.38 (C-9), 56.43 (3-OCH3), 35.87 (C-8), 32.7 (C-7).
1,2-Dihydrodendroarboreol B (12) − Light yellow oil; C17H24O2; [a]20D = -29° (c 0.04, MeOH); ESI-Q-TOF-MS: m/z 374.1741 [M+Na]+; 1H-NMR (CD3OD, 500 MHz): δ 6.30 (1H, dd, J = 15.9, 5.5 Hz, H-9), 5.81 (1H, m, H-16), 5.76 (1H, ddd, J = 15.9, 1.7, 0.8 Hz, H-8), 4.99 (1H, dq, J = 17.1, 1.5 Hz, H-17a), 4.92 (1H, ddt, J = 10.2, 2.3, 1.5 Hz, H-17b), 4.33 (1H, t, J = 6.6 Hz, H-3), 4.10 (1H, q, J = 5.5 Hz, H-10), 2.05 (2H, m, H-15), 1.69 (2H, m, H-2), 1.49 (2H, m, H-11), 1.38-1.28 (6H, m, H-12, 13, 14), 1.00 (3H, t, J = 7.4 Hz, H-1); 13C-NMR (CD3OD, 125 MHz): δ 151.70 (C-9), 140.20 (C-16), 114.93 (C-17), 108.69 (C-8), 84.56 (C-4), 77.51 (C-7), 74.38 (C-6), 72.61 (C-10), 69.62 (C-5), 64.65 (C-3), 37.95 (C-11), 34.96 (C-15), 31.95 (C-2), 30.23 (C-13), 30.17 (C-14), 26.43 (C-12), 9.97 (C-1).
1,2-Dihydropanaxydiol (13) − Colorless oil; C17H26O2; [a]25D = +342.70° (c 0.037, CHCl3); ESI-Q-TOF-MS: m/z 374.1741 [M+Na]+; 1H-NMR (CD3OD, 500 MHz): δ 6.30 (1H, dd, J = 15.9, 5.6 Hz, H-9), 5.76 (1H, ddd, J = 15.9, 1.7, 0.8 Hz, H-8), 4.33 (1H, t, J = 6.6 Hz, H-3), 4.10 (1H, m, H-10), 1.69 (2H, m, H-2), 1.49 (2H, q, J = 6.8 Hz, H-11), 1.38-1.23 (10H, m, H-12, 13, 14, 15, 16), 0.90 (3H, m, H-17); 13C-NMR (CD3OD, 125 MHz): δ 151.73 (C-9), 108.67 (C-8), 84.56 (C-4), 77.52 (C-7), 74.37 (C-6), 72.65 (C-10), 69.62 (C-5), 64.65 (C-3), 38.02 (C-11), 33.14 (C-15), 31.96 (C-2), 30.77 (C-13), 30.52 (C-14), 26.61 (C-12), 23.85 (C-16), 14.56 (C-17), 9.96 (C-1).
Panaxydiol (14) − Yellowish oil; C17H24O2; [a]20D = -33.6° (c 0.05, CHCl3); ESI-Q-TOF-MS: m/z 374.1741 [M+Na]+; 1H-NMR (CD3OD, 500 MHz): δ 6.32 (1H, dd, J = 15.9, 5.6 Hz, H-9), 5.92 (1H, ddd, J = 17.1, 10.2, 5.5 Hz, H-2), 5.77 (1H, dt, J = 15.9, 1.2 Hz, H-8), 5.40 (1H, dt, J = 17.1, 1.4 Hz, H-1a), 5.20 (1H, dt, J = 10.2, 1.4 Hz, H-1b), 4.90 (1H, m, H-3), 4.10 (1H, m, H-10), 1.49 (2H, m, H-11), 1.37-1.24 (10H, m, H-12, 13, 14, 15, 16), 1.33 (3H, m, H-17); 13C-NMR (CD3OD, 125 MHz): δ 152.01 (C-9), 138.26 (C-2), 116.73 (C-1), 108.52 (C-8), 82.35 (C-4), 78.14 (C-7), 74.18 (C-6), 72.61 (C-10), 70.74 (C-5), 64.14 (C-3), 38.00 (C-11), 33.13 (C-15), 30.76 (C-13), 30.52 (C-14), 26.61 (C-12), 23.84 (C-16), 14.57 (C-17).
Panaxytriol (15) − Colorless needles; C17H26O3; [a]20D = -12.0° (c 0.6, CHCl3); ESI-Q-TOF-MS: m/z 301.1773 [M+Na]+; 1H-NMR (CD3OD, 500 MHz): δ 5.90 (1H, ddd, J = 17.1, 10.1, 5.5 Hz, H-2), 5.39 (1H, dt, J = 17.1, 1.4 Hz, H-1a), 5.18 (1H, dt, J = 10.1, 1.4 Hz, H-1b), 4.85 (1H, dt, J = 5.5, 1.3 Hz, H-3), 3.59 (1H, ddd, J = 6.9, 6.0, 3.5 Hz, H-9), 3.53 (1H, dt, J = 7.8, 3.5 Hz, H-10), 2.58 (1H, ddd, J = 17.3, 6.0, 1.0 Hz, H-8a), 2.48 (1H, ddd, J = 17.3, 6.9, 1.0 Hz, H-8b), 1.24-1.53 (12H, m, H-12, 13, 14, 15, 16), 0.91 (3H, m, H-17); 13C-NMR (CD3OD, 125 MHz): δ 138.42 (C-2), 116.60 (C-1), 79.80 (C-7), 76.08 (C-4), 73.88 (C-10), 73.62 (C-9), 71.22 (C-5), 66.61 (C-6), 63.99 (C-3), 34.16 (C-11), 33.20 (C-15), 30.89 (C-13), 30.60 (C-14), 27.17 (C-8), 25.02 (C-12), 23.89 (C-16), 14.62 (C-7).
(S)-cis-Abscisic acid (16) − Light yellow oil; C15H20O4; [a]25D = +303.33° (c 0.03, MeOH); CD (MeOH) : Δε (nm) = -1.23 (321), +20.9 (264), -15.8 (232); ESI-Q-TOF-MS: m/z 287.1257 [M+Na]+; 1H-NMR (CD3OD, 500 MHz): δ 7.79 (1H, d, J = 16.5 Hz, H-8), 6.24 (1H, d, J = 16.5 Hz, H-7), 5.93 (1H, m, H-4), 5.75 (1H, s, H-10), 2.54 (1H, d, J = 17.2 Hz, H-2a), 2.18 (1H, d, J = 17.2 Hz, H-2b), 2.04 (3H, d, J = 1.3 Hz, H-15), 1.93 (3H, d, J = 1.3 Hz, H-14), 1.07 (3H, s, H-12), 1.03 (3H, s, H-13); 13C-NMR (125 MHz, CD3OD): δ 201.18 (C-3), 166.69 (C-5), 151.31 (C-9), 138.13 (C-7), 129.53 (C-8), 127.72 (C-4), 119.66 (C-10), 80.74 (C-6), 50.79 (C-2), 43.00 (C-1), 24.80 (C-13), 23.70 (C-12), 21.40 (C-15), 19.76 (C-14).
(R)-trans-Abscisic acid (17) − Light yellow oil; C15H20O4; [a]25D = +238.25° (c 0.1, MeOH); CD (MeOH): Δε (nm) = -2.1 (320), +28.3 (268), -24.7 (233); ESI-Q-TOF-MS: m/z 287.1256 [M+Na]+; 1H-NMR (CD3OD, 500 MHz): δ 6.49 (1H, d, J = 15.7 Hz, H-8), 6.28 (1H, d, J = 15.7 Hz, 1H, H-7), 5.91 (1H, p, J = 1.3 Hz, H-4), 5.84 (1H, s, H-10), 2.56 (1H, d, J = 17.0 Hz, H-2a), 2.28 (3H, d, J = 1.3 Hz, H-15), 2.23 (1H d, J = 17.0 Hz, H-2b), 1.91 (3H, d, J = 1.3 Hz, H-14), 1.06 (3H, s, H-12), 1.01 (3H, s, H-13); 13C-NMR (CD3OD, 125 MHz): δ 201.05 (C-3), 170.39 (C-11), 166.41 (C-5), 152.71 (C-9), 137.09 (C-7), 135.31 (C-8), 127.59 (C-2), 121.47 (C-10), 80.46 (C-6), 50.81 (C-2), 42.91 (C-1), 24.85 (C-13), 23.72 (C-12), 19.54 (C-14), 14.28 (C-15).
(6R,9S)-Blumenol C (18) − Colorless gum; C13H22O2; [α]25D = +10.17° (c 0.12, MeOH); CD (MeOH): Δε (nm) = +0.47 (332), -0.12 (264), +1.81 (212); ESI-Q-TOF-MS: m/z 211.1694 [M+H]+; 1H-NMR (CD3OD, 500 MHz): δ 5.81 (1H, t, J = 1.2 Hz, H-4), 3.69 (1H, m, J = 6.2 Hz, H-9), 2.45 (1H, d, J = 17.4 Hz, H-2a), 2.04 (3H, d, J = 1.3 Hz, H-11), 1.99 (2H, m, H-2b, 6), 1.76 (1H, m, H-7b), 1.62 (1H, m, H-7a), 1.53 (2H, m, H-8), 1.17 (3H, d, J = 6.2 Hz, H-10), 1.09 (3H, d, J = 2.2 Hz, H-13), 1.02 (3H, s, H-12); 13C-NMR (CD3OD, 125 MHz): δ 202.40 (C-3), 169.93 (C-5), 125.58 (C-4), 69.00 (C-9), 52.56 (C-6), 48.22 (C-2), 39.98 (C-8), 37.47 (C-1), 29.16 (C-12), 27.60 (C-7), 27.53 (C-13), 25.04 (C-11), 23.66 (C-10).
(3S,5R,6S,7E)-5,6-Epoxy-3-hydroxy-7-megastigmene-9-one (19) − Amorphous powder; C13H20O3; [a]25D = -112.3° (c 0.15, MeOH); CD (MeOH): Δε (nm) = -8.11 (233); ESI-Q-TOF-MS: m/z 225.1483 [M+H]+; 1H-NMR (CD3OD, 500 MHz): δ 7.19 (1H, d, J = 15.8 Hz, H-7), 6.19 (1H, d, J = 15.8 Hz, H-8), 3.76 (1H, dddd, J = 10.8, 9.0, 4.9, 3.4 Hz, H-3), 2.29 (5H, m, H-4a, 10), 1.65 (1H, dd, J = 14.3, 9.2 Hz, H-4b), 1.58 (1H, ddd, J = 12.9, 3.4, 1.8 Hz, H-2a), 1.27 (1H, m, H-2b), 1.19 (3H, s, H-11), 1.18 (3H, s, H-13), 0.96 (3H, s, H-12); 13C-NMR (CD3OD, 125 MHz): δ 200.39 (C-9), 145.59 (C-7), 133.96 (C-8), 70.98 (C-6), 68.94 (C-5), 64.52 (C-3), 47.87 (C-2), 41.47 (C-4), 36.24 (C-1), 29.91 (C-11), 27.57 (C-10), 25.27 (C-12), 20.18 (C-13).
(+)-Salicifoliol (20) − Colorless oil; C13H14O5; [a]25D = 56.33° (c 0.06, MeOH); CD (MeOD): Δε (nm) = +0.08 (285), +2.31 (211), +0.40 (202); ESI-Q-TOF-MS: m/z 273.0738 [M+Na]+; 1H-NMR (CDCl3, 500 MHz): δ 6.89 (1H, d, J = 8.1 Hz, H-5'), 6.86 (1H, d, J = 2.0 Hz, H-2'), 6.79 (1H, dd, J = 8.1, 2.0 Hz, H-6'), 4.59 (1H, d, J = 7.0 Hz, H-6), 4.48 (1H, dd, J = 9.8, 6.8 Hz, H-4a), 4.35 (1H, t, J = 9.1 Hz, H-8a), 4.31 (1H, dd, J = 9.8, 2.1 Hz, H-4b), 4.17 (1H, dd, J = 9.1, 3.9 Hz, H-8b), 3.89 (3H, s, 3-OCH3), 3.43 (1H, td, J = 9.0, 3.9 Hz, H-1), 3.10 (1H, m, H-5); 13C-NMR (CDCl3, 125 MHz): δ 178.45 (C-2), 147.11 (C-3'), 146.04 (C-4'), 130.76 (C-1'), 119.34 (C-6'), 114.62 (C-5'), 108.61 (C-2'), 86.30 (C-6), 70.19 (C-8), 70.04 (C-4), 56.19 (3-OCH3), 49.41 (C-5), 46.23 (C-1).
Imperatorin (21) − Colorless crystalline solid; C16H14O4; ESI-Q-TOF-MS: m/z 271.097 [M+H]+; 1H-NMR (CDCl3, 500 MHz): δ 7.75 (1H, d, J = 9.5 Hz, H-4), 7.67 (1H, d, J = 2.2 Hz, H-2'), 7.34 (1H, s, H-5), 6.80 (1H, d, J = 2.2 Hz, H-3'), 6.35 (1H, d, J = 9.5 Hz, H-3), 5.59 (1H, t, J = 7.2 Hz, H-2"), 4.99 (2H, d, J = 7.2 Hz, H-1"), 1.72 (3H, d, J = 1.4 Hz, H-4"), 1.70 (3H, d, J = 1.4 Hz, H-5"); 13C-NMR (CDCl3, 125 MHz): δ 160.82 (C-2), 148.82 (C-7), 146.85 (C-2'), 144.61 (C-4), 144.02 (C-9), 140.04 (C-3"), 131.88 (C-8), 126.08 (C-6), 119.95 (C-2"), 114.02 (C-9), 114.92 (C-5), 113.37 (C-3), 106.93 (C-3'), 70.38 (C-1"), 25.07 (C-4"), 18.36 (C-5").
Indole-3-carboxaldehyde (22) − Colorless solid; C9H7NO; ESI-Q-TOF-MS: m/z 144.0449 [M-H]-; 1H-NMR (CD3OD, 500 MHz): δ 9.89 (1H, s, 3-COH), 8.16 (1H, dd, J = 7.3, 1.6 Hz, H-4), 8.11 (1H, s, H-2), 7.48 (1H, dd, J = 7.9, 1.2 Hz, H-7), 7.26 (2H, m, H-5, 6); 13C-NMR (CD3OD, 125 MHz): δ 187.57 (3-COH), 139.90 (C-2), 139.09 (C-9), 125.86 (C-8), 125.16 (C-6), 123.77 (C-5), 122.55 (C-4), 120.28 (C-3), 113.28 (C-7).
Computational methods − The 3D models of the four diastereomers (3S,6S; 3R,6S; 3R,6R; 3S,6R) of 1 were generated using Chem3D 15.0 (PerkinElmer). Conformational searches were then conducted in the Spartan 14 software (Wavefunction Inc.) using the Merck Molecular Force Field (MMFF) within 10 kJ/mol energy window. The optimization of selected conformers was performed by density functional theory (DFT) calculations at the B3LYP/6–31+G(d,p) in chloroform using the Gaussian 09 package (Wallingford, CT, USA). Time-dependent density functional theory (TDDFT) calculations were carried out at the CAM-B3LYP/6–31+G(d,p) level in chloroform with conductor-like polarizable continuum model (CPCM) model. The calculated ECD curves were visualized using SpecDis v1.71 software (Berlin, Germany).
Results and Discussion
By comparing the 1H, 13C-NMR, and MS spectroscopic data with relevant references, the chemical structures of compounds 1−22 were identified. The known compounds (2–22) were identified to be dihydrosinapic acid (2),17 2,6-dimethoxyterephthalic acid (3),18 4-methoxybenzoic acid (4),19 4-ethylbenzene-1,2-diol (5),20 ethylparaben (6),21 vanillic acid (7),22 ferulic acid (8),23 3-(4-hydroxy-3-methoxyphenyl)propionic acid (9),24 ethyl dihydroferulate (10),25 dihydroconiferyl alcohol (11),26 1,2-dihydrodendroarboreol B (12),27 1,2-dihydropanaxydiol (13),28 panaxydiol (14),28 panaxytriol (15),29 (S)-cis-abscisic acid (16),30 (R)-trans-abscisic acid (17),30 (6R,9S)-blumenol C (18),31 (3S,5R,6S,7E)-5,6-epoxy-3-hydroxy-7-megastigmene-9-one (19),32 (+)-salicifoliol (20),33 imperatorin (21),34 and indole-3-carboxaldehyde (22) (Fig. 1).35
Compound 1 was isolated as a white powder. Its molecular formula, C13H20O3, was established based on the [M-H]- ion peak at m/z 223.1332 (calculated for C13H19O3, 223.1334) observed in the ESI-Q-TOF-MS analysis. In the 1H-NMR spectrum, three methyl groups were observed at δH 1.65 (3H, s, H-10), 1.06 (3H, s, H-13), and 1.03 (3H, s, H-12), along with three pair of methylene signals at δH 2.44 – 2.24 (2H, m, H-7a, 8a), 2.04 (1H, m, H-8b), 1.94 (1H, ddd, J = 13.3, 6.4, 1.7 Hz, H-2a), 1.87 (1H, ddd, J = 12.7, 9.3, 5.0 Hz, H-7b), and 1.51 (1H, dd, J = 13.3, 9.8 Hz, H-2b). Moreover, the spectrum showed signals for one olefinic proton [δH 6.88 (1H, dd, J = 2.7, 1.7 Hz, H-4)] and one methine with a hydroxy group [δH 4.43 (1H, ddd, J = 9.8, 6.4, 2.7 Hz, H-3)]. In the 13C-NMR spectrum, a total of 13 signals were observed, including one quaternary carbon, one oxygenated quaternary carbon, four olefinic carbons, three methylene carbons, three methyl carbons, and one methine carbon, with reliability supported by the HSQC spectrum (Fig. 2 and Supplementary information S1–S6).
The HMBC spectrum displayed correlation peaks between H-2 and C-1/C-3/C-4/C-6; H-3 and C-5; H-4 and C-5/C-6/C-11; H-7 and C-5/C-6/C-8/C-9; H-8 and C-6/C-7/C-9/C-10; H-10 and C-9/C-11; H-12 and C-1/C-6/C-13; H-13 and C-1/C-2/C-12 (Fig. 2B), which indicated that compound 1 had a bicyclic megastigmane skeleton (Fig. 2 and Supplementary information S7).
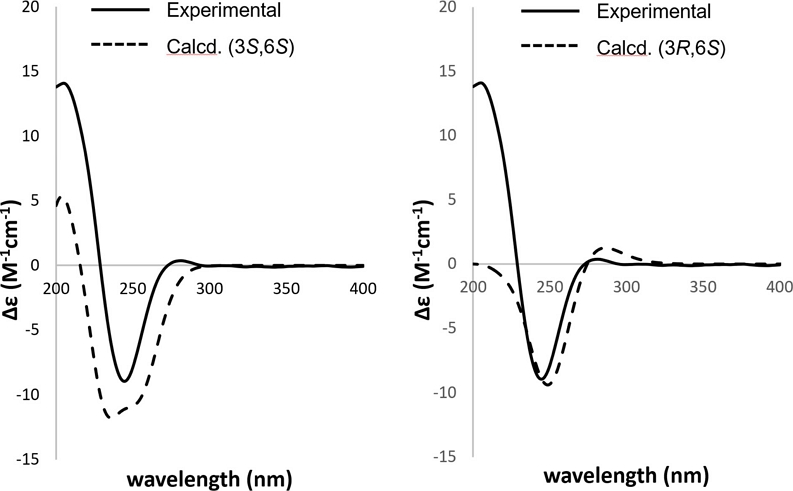
The absolute configurations of C-3 and C-6 of 1 were determined through a comparison between the experimental and calculated ECD spectra. Among the four conformers, two pairs of enantiomers, specifically (3S,6R) and (3R,6S), as well as (3R,6R) and (3S,6S), showed opposite patterns in the ECD spectrum. When comparing the Cotton effects of the two conformers, (3R,6S) and (3S,6S), the most significant difference observed was that only (3S,6S) closely resembles the experimental data, exhibiting a similar positive Cotton effect at 205 nm. Furthermore, the similarity factor for the (3S,6S) form of 1 was estimated as 0.8963 (σ = 0.31 eV; shift = 6 nm) using SpecDis v1.71 software (Fig. 3, Supplementary Information S8–S9). Based on the above spectroscopic data, the chemical structure of 1 was determined as shown in Fig. 1 with a trivial name of (3S,6S)-amoreol, and its presence in nature is reported here for the first time.
Compound 1 possesses the C13 megastigmane bicyclic norsesquiterpene skeleton. This structure arises from the oxidative degradation of β-carotenoids biosynthesized through the isoprene pathway, where the numerous double bonds within carotenoids give rise to a variety of apocarotenoid derivatives.36,37 Particularly, bicyclic megastigmane compound is rarely found.38
Continuous research has been dedicated to discovering the bioactive constituents from P. ginseng for a long time and is still being extensively conducted. In the industry that utilizes P. ginseng, the disposing of the residual by-products, excluding the primary active compounds, is a highly detrimental process from an industrial standpoint. Therefore, attributing value to the by-products through chemical analysis or biological evaluation studies on their activities and reutilizing them can be beneficial in various ways.
Acknowledgments
This work was supported by research fund from National Research Foundation of Korea (# NRF-2018R1A6A1A03025108) and Amorepacific Corporation (2022).
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
-
Lee, D. Y.; Kim, H. G.; Lee, Y. G.; Kim, J. H.; Lee, J. W.; Choi, B. R.; Jang, I. B.; Kim, G. S.; Baek, N. I. Molecules 2018, 23, 267.
[https://doi.org/10.3390/molecules23020267]

-
Yun, T. K. J. Korean Med Sci. 2001, 16 Suppl, S3-S5.
[https://doi.org/10.3346/jkms.2001.16.S.S3]

-
So, S. H.; Lee, J. W.; Kim, Y. S.; Hyun, S. H.; Han, C. K. J. Ginseng Res. 2018, 42, 549-561.
[https://doi.org/10.1016/j.jgr.2018.05.002]

-
Cho, J. H.; Song, M. C.; Lee, Y.; Noh, S. T.; Kim, D. O.; Rha, C. S. J. Ginseng Res. 2023, 47, 593-603.
[https://doi.org/10.1016/j.jgr.2023.02.006]

-
Zhang, S.; Ding, C.; Liu, X.; Zhao, Y.; Ding, Q.; Sun, S.; Zhang, J.; Yang, J.; Liu, W.; Li, W. Molecules 2023, 28, 3733.
[https://doi.org/10.3390/molecules28093733]

-
Lee, E. J.; Shaykhutdinov, R.; Weljie, A. M.; Vogel, H. J.; Facchini, P. J.; Park, S. U.; Kim, Y. K.; Yang, T. J. J. Agric. Food Chem. 2009, 57, 7513-7522.
[https://doi.org/10.1021/jf901675y]

-
Matsunaga, H.; Katano, M.; Yamamoto, H.; Mori, M.; Takata, K. Chem. Pharm. Bull (Tokyo). 1989, 37, 1279-1281.
[https://doi.org/10.1248/cpb.37.1279]

-
Choi, K. T. Acta Pharmacol. Sin. 2008, 29, 1109-1118.
[https://doi.org/10.1111/j.1745-7254.2008.00869.x]

-
Wang, D.; Shao, S.; Zhang, Y.; Zhao, D.; Wang, M. Front. Immunol. 2021, 12, 683911.
[https://doi.org/10.3389/fimmu.2021.683911]

-
Zhou, S. S.; Xu, J.; Zhu, H.; Wu, J.; Xu, J. D.; Yan, R.; Li, X. Y.; Liu, H. H.; Duan, S. M.; Wang, Z.; Chen, H. B.; Shen, H.; Li, S. L. Sci. Rep. 2016, 6, 22474.
[https://doi.org/10.1038/srep22474]

-
Fan, J. W.; Xu, X. T.; Cheng, H.; Sang, Z.; Shi, Y. H. Am. J. Chin. Med. 2023, 51, 909-927.
[https://doi.org/10.1142/S0192415X23500428]

- Hu, J. R.; Chun, Y. S.; Kim, J. K.; Cho, I. J.; Ku, S. K. Exp. Ther. Med. 2019, 18, 4388-4396.
-
Attele, A. S.; Zhou, Y. P.; Xie, J. T.; Wu, J. A.; Zhang, L.; Dey, L.; Pugh, W.; Rue, P. A.; Polonsky, K. S.; Yuan, C. S. Diabetes 2002, 51, 1851-1858.
[https://doi.org/10.2337/diabetes.51.6.1851]

-
Kim, S. W.; Gupta, R.; Lee, S. H.; Min, C. W.; Agrawal, G. K.; Rakwal, R.; Kim, J. B.; Jo, I. H.; Park, S. Y.; Kim, J. K.; Kim, Y. C.; Bang, K. H.; Kim, S. T. Front. Plant Sci. 2016, 7, 994.
[https://doi.org/10.3389/fpls.2016.00994]

- Jin, S.; Eom, S. H.; Kim, J. S.; Jo, I. H.; Hyun, T. K. J. Appl. Bot. Food Qual. 2019, 92, 130–137.
-
Kim, M. S.; Kim, S. H.; Park, S. J.; Sung, M. J.; Park, J.; Hwang, J. T.; Yang, H. J.; Kim, S.; Seo, D.; Shin, S. S.; Hur, H. J. J. Funct. Foods 2017, 35, 295-302.
[https://doi.org/10.1016/j.jff.2017.05.050]

-
Takahashi, T.; Miyazawa, M. Pharmazie 2010, 65, 913-918.
[https://doi.org/10.2524/jtappij.63.913]

-
Nishanbaev, S. Z.; Bobakulov, K. M.; Abdullaev, N. D.; Sham’yanov, I. D. Chem. Nat. Compd. 2015, 51, 537-539.
[https://doi.org/10.1007/s10600-015-1334-4]

-
Zhang, X.; Zhang, W.-Z.; Shi, L.-L.; Guo, C.-X.; Zhang, L.-L.; Lu, X.-B. Chem. Commun (Camb). 2012, 48, 6292-6294.
[https://doi.org/10.1039/c2cc32045b]

-
Bomon, J.; Bal, M.; Achar, T. K.; Sergeyev, S.; Wu, X.; Wambacq, B.; Lemière, F.; Sels, B. F.; Maes, B. U. W. Green Chem. 2021, 23, 1995-2009.
[https://doi.org/10.1039/D0GC04268D]

-
Liu, L.; Li, Z.; Chen, C.; Li, H.; Xu, L.; Yu, Z. Tetrahedron 2018, 74, 2447-2453.
[https://doi.org/10.1016/j.tet.2018.03.070]

-
Sun, J.; Zhang, F.; Yang, M.; Zhang, J.; Chen, L.; Zhan, R.; Li, L.; Chen, Y. Nat. Prod. Res. 2014, 28, 1900-1905.
[https://doi.org/10.1080/14786419.2014.955495]

-
Elbermawi, A.; Halim, A. F.; Mansour, E. S.; Ahmad, K. F.; Elsbaey, M.; Ashour, A.; Amen, Y.; El-Gamil, M. M.; Tomofumi, M.; Shimizu, K. Nat. Prod. Res. 2022, 36, 5134-5141.
[https://doi.org/10.1080/14786419.2021.1922902]

-
Zhu, M.; Shan, Q.; Ma, L.; Wen, J. J.; Dong, B. A.; Zhang, G. N.; Wang, M. H.; Wang, J. X.; Zhou, J. M.; Cen, S.; Wang, Y. C. Eur. J. Med. Chem. 2021, 220, 113498.
[https://doi.org/10.1016/j.ejmech.2021.113498]

-
Ishimata, N.; Ito, H.; Tai, A. Bioorg. Med. Chem. Lett. 2016, 26, 3533-3536.
[https://doi.org/10.1016/j.bmcl.2016.06.028]

-
Huang, Y. H.; Zeng, W. M.; Li, G. Y.; Liu, G. Q.; Zhao, D. D.; Wang, J.; Zhang, Y. L. Molecules 2014, 19, 507-513.
[https://doi.org/10.3390/molecules19010507]

-
Jiang, M.-Y.; Yang, C.-T.; Pu, X.-Y.; Fu, G.; Wang, W.; Li, Y.-X.; Feng, L.; Niu, H.-R.; Tan, J.-L.; Huang, X.-Z. Rec. Nat. Prod. 2019, 13, 424-428.
[https://doi.org/10.25135/rnp.119.18.09.940]

-
Resetar, M.; Liu, X.; Herdlinger, S.; Kunert, O.; Pferschy-Wenzig, E. M.; Latkolik, S.; Steinacher, T.; Schuster, D.; Bauer, R.; Dirsch, V. M. J. Nat. Prod. 2020, 83, 918-926.
[https://doi.org/10.1021/acs.jnatprod.9b00691]

-
Yun, H. D.; Danishefsky, S. J. J. Org. Chem. 2003, 68, 4519-4522.
[https://doi.org/10.1021/jo0341665]

-
Ferreres, F.; Andrade, P.; Tomás-Barberán, F. A. J. Agric. Food Chem. 1996, 44, 2053-2056.
[https://doi.org/10.1021/jf9507553]

-
Yan, J.; Shi, X.; Donkor, P. O.; Zhu, H.; Gao, X.; Ding, L.; Qiu, F. J. Nat. Med. 2017, 71, 780-790.
[https://doi.org/10.1007/s11418-017-1102-9]

-
D'Abrosca, B.; DellaGreca, M.; Fiorentino, A.; Monaco, P.; Oriano, P.; Temussi, F. Phytochemistry 2004, 65, 497-505.
[https://doi.org/10.1016/j.phytochem.2003.11.018]

-
Zhou, L.; Han, F. Y.; Lu, L. W.; Yao, G. D.; Zhang, Y. Y.; Wang, X. B.; Lin, B.; Huang, X. X.; Song, S. J. Phytochemistry 2019, 164, 122-129.
[https://doi.org/10.1016/j.phytochem.2019.05.008]

-
Tavakoli, S.; Delnavazi, M. R.; Hadjiaghaee, R.; Jafari-Nodooshan, S.; Khalighi-Sigaroodi, F.; Akhbari, M.; Hadjiakhoondi, A.; Yassa, N. Nat. Prod. Res. 2018, 32, 2724-2728.
[https://doi.org/10.1080/14786419.2017.1375915]

-
Liao, J. H.; Yuan, C. M.; Di, Y. T.; He, H. P.; Hu, X. J. Asian J. Chem. 2014, 26, 4504-4506.
[https://doi.org/10.14233/ajchem.2014.17486]

-
Kornpointner, C.; Hochenegger, N. J.; Shi, B. B.; Berger, A.; Theiner, J.; Brecker, L.; Schinnerl, J. Molecules 2022, 27, 7284.
[https://doi.org/10.3390/molecules27217284]

-
Rao, A. S. Chem. Int. 2017, 3, 69-91.
[https://doi.org/10.1007/BF03400310]

-
Yaermaimaiti, S.; Turak, A.; Huang, Q.; Liu, G.; Zhao, J.; Aisa, H. A. Phytochemistry 2022, 203, 113361.
[https://doi.org/10.1016/j.phytochem.2022.113361]