A New Ring-opened Azaphilone from a Fungus Talaromyces sp. FB15
Abstract
Eleven compounds (1–11), including a new azaphilone derivative (1), were isolated from cultures of Talaromyces sp. FB15, a marine fungus collected in the Beaufort Sea in northern Alaska. The structures of the isolated compounds were established by analysis of 1D and 2D NMR data, and they all were turned out to be polyketides such as azaphilone, anthraquinone, and anthrones. Interestingly, the structure of the new metabolite (1) was determined to have an azaphilone scaffold with the pyran ring opened, and to our knowledge, this is the second report on the isolation of this type of azaphilones.
Keywords:
Marine fungus, Polyketides, Azaphilone, Ring-opened azaphiloneIntroduction
Ninety-five percent of the Earth’s ecosystem and more than 70% of the planet’s surface are found in the water. Fungi originating from the sea produce distinct metabolites that differ structurally from those of terrestrial fungi. As a result, since the 1970s, much research has been carried out on the secondary metabolites of marine mushrooms or fungi derived from the sea. Specifically, studies conducted between 2005 and 2017 demonstrate the biological potential of marine-derived fungi as well as the structural distinctiveness of the secondary metabolites they create. Furthermore, it is evident that the majority of studies on fungus originating from the sea have been carried out using marine sediments that have been obtained from mangrove areas, deep sea, and shallow seas.1 Up to 1,000 meters below the surface of the ocean, deep-sea fungus thrive. It is generally understood to be a type of fungus that thrives in a particular habitat devoid of sunlight, high water pressure, and low temperatures. In addition to producing secondary metabolites that have therapeutic uses, deep-sea isolated fungi are particularly well-known for their chemical and biological variety. A review report states that to date, over 180 secondary metabolites exhibiting biological action have been identified and isolated.2–3
As a part of a study on the secondary metabolites produced by marine fungi, some fungal strains were obtained from marine sediments collected from the Beaufort Sea in northern Alaska. Due to the unique HPLC-UV profiles of the culture extracts, a strain FB15 was selected for chemical investigation. Utilizing the ITS sequencing approach, it was determined that the identity of this strain corresponded to Talaromyces sp. Following the extraction of the strain’s large-scale cultures in an organic solvent, eleven secondary metabolites, including a new compound, were isolated using a variety of chromatographic techniques. The majority of the isolated chemicals were polyketides, including anthrones, azaphilones, and anthraquinones.
Experimental
General experimental procedures – NMR spectra were recorded on a Bruker DPX400 spectrometer (400 MHz for 1H and 100 MHz for 13C), using TMS as an internal standard. ESIMS spectra were obtained from Agilent Technologies 6130 mass spectrometer using a C18 (Merck, Lichroprep RP-18) column. HRESIMS spectra were obtained from a Waters UHPLC Xevo G2 Q-TOF. Preparative MPLC was performed on a Biotage Selekt using silica gel (Sfär Silica) and C18 (Sfär C18). Preparative HPLC was performed on a Waters Millipore 600 system using a C18 (Phenomenex, Luna 5 μm C18 (2) 100 Å, 250 × 21.2 mm) column. Semi-preparative HPLC was performed on a Waters Millipore 600 system using a C18 (YMC 5 μm C18, 250 × 10 mm) column. UV spectra were detected on PDA 996. High-resolution electrospray ionization (ESI) mass spectra were obtained using a micromass ZQ (Waters, Manchester, UK) and a UHR ESI Q-TOF (Bruker, Billerica, MA, USA) mass spectrometer. All solvent used for experiments were of analytical grade solvents.
Fungal materials – The fungal strain Talaromyces sp. FB15, which was isolated from marine sediment collected in the Beaufort Sea in northern Alaska, was provided by Emeritus professor Jong Heon Shin. The strain was identified by ITS sequencing method. A voucher specimen was deposited at the Natural Products Chemistry laboratory of College of Pharmacy, Seoul National University, Seoul, South Korea.
Extraction and isolation – The strain FB15 was cultivated on PDA media. A 1 L PDA medium was made in 11 culture flasks at a ratio of 12 g potato dextrose broth and 11.8 g agar in 500 mL of water. The prepared medium was autoclaved at 125°C for about 20 minutes. The sterilized medium was cooled and coagulated in a clean bench for about a day. The seed was inoculated into a PDA medium in a clean bench. After incubating for 3 weeks, the medium in which the fungi were cultured was crushed with a spoon, and extracted using ethyl acetate. The extraction was carried out three times at intervals of one day, which afforded 4.2 g of the extracts. The crude extract was subjected to medium pressure liquid chromatography (MPLC) on silica gel (300 g, Sfär Silica) using a gradient phase of chloroform and methanol, to afford 22 fractions (Fr.1–22). Fraction 9 was purified by reversed-phase HPLC (Phenomenex Luna C18 (2), 5 μm, 250 × 10.0 mm, 2.0 mL/min, isocratic of 70% CH3CN in H2O, UV 210, 254, 280 and 365 nm) over C18 to afford compound 1 (1.0 mg). Compounds 2 (2.6 mg), 3 (1.5 mg), 4 (8.1 mg), and 5 (4.6 mg) were isolated from fraction 10 by reversed-phase HPLC (Phenomenex Luna C18 (2), 5 μm, 250 × 10.0 mm, 2.0 mL/min, isocratic of 75% CH3CN in H2O, UV 210, 254, 280 and 365 nm). Fraction 11 was subjected to reversed-phase HPLC (Phenomenex Luna C18 (2), 5 μm, 250 × 10.0 mm, 5.0 mL/min, isocratic of 80% CH3CN in H2O, UV 210, 254, 280 and 365 nm) to afford compound 6 (2.0 mg). Compounds 7 (1.2 mg) and 8 (2.3 mg) were isolated from fraction 13 using reversed-phase semi-preparative HPLC (YMC 5 μm C18, 250 × 10 mm; flow 2 mL/min; isocratic 24% CH3CN in H2O, UV 210, 254, 280 and 365 nm). Fraction 14 was separated using MPLC by elution of CH3CN/H2O (1/9, 3/7, 1/1, 7/3, 4/1, 9/1, 10/0) to afford seven subfractions (Fr.14-1–7). The subfraction 14-2 was further subjected to reversed-phase semi-preparative HPLC (YMC 5 μm C18, 250 × 10 mm; flow 2 mL/min; isocratic 24% CH3CN in H2O, UV 210, 254, 280 and 365 nm) leading to the isolation of compounds 9 (1.2 mg) and 10 (13.5 mg). Fraction 18 was also separated using MPLC (5 g, Sfär C18) by elution of CH3CN/H2O (1/9, 3/7, 1/1, 7/3, 4/1, 9/1, 10/0) to afford six subfractions (Fr.18-1–6). The subfraction 18-1 was further subjected to reversed-phase preparative HPLC (Phenomenex Luna 5 μm C18, 250 × 21.2 mm; flow rate, 5 mL/min; isocratic 50% CH3CN in H2O, UV 210, 254, 280 and 365 nm) to afford compound 11 (14 mg).
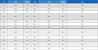
Methyl-talarophilone (1) – Yellow amorphous solid; 1H and 13C NMR, see Table 1.; (+)HRESIMS m/z 404.1823 [M]+, calcd for C22H28O7 at m/z 404.1835.
4-Hydroxyemodin (2) –White amorphous solid; 1H NMR (400 MHz, CD3OD): δ 7.32 (d, J = 1.1 Hz), 7.06 (s), 6.66 (s), 2.35 (s); (+) ESIMS m/z 287.1 [M+H]+, calcd for C15H11O6 at m/z 287.0550.
Kasanosin C (3) – Yellow amorphous solid; 1H NMR (400 MHz, CD3OD): δ 6.46 (m), 6.24 (d, J = 2.0 Hz), 6.20 (d, J = 2.0 Hz), 6.02 (dd, J = 15.0, 1.6 Hz), 5.44 (s), 4.86 (d, J = 12.4 Hz), 4.80 (d, J = 12.4 Hz), 4.00 (dd, J = 8.0, 4.8 Hz), 3.68 (s), 2.81 (dd, J = 17.5, 6.6 Hz), 2.60 (dd, J = 17.5, 5.0 Hz), 2.16 (s), 1.85 (dd, J = 7.0, 1.5 Hz), 1.67 (s); (+) ESIMS m/z 401.2 [M+H]+, calcd for C22H25O7 at m/z 401.1595.
Monomethyl-(S)-mitorubrin (4) – Red amorphous solid; = −20.8; 1H NMR (400 MHz, CD3OD): δ 8.15 (s), 6.66 (m), 6.45 (s), 6.30 (d, J = 1.5 Hz), 6.25 (d, J = 1.5 Hz), 6.19 (dd, J = 15.0, 1.5 Hz), 5.63 (d, J = 1.0 Hz), 3.76 (s), 2.40 (s), 1.94 (dd, J = 7.0, 1.5 Hz), 1.56 (s); (+) ESIMS m/z 397.2 [M+H]+, calcd for C22H21O7 at m/z 397.1282.
Comazaphilone A (5) – Yellow amorphous powder; 1H NMR (400 MHz, CDCl3): δ 6.18 (d, J = 1.5 Hz), 6.16 (d, J = 1.5 Hz), 5.45 (t, J = 3.0 Hz), 5.13 (s), 4.98 (d, J = 12.6 Hz), 4.76 (d, J = 12.6 Hz), 3.62 (s), 2.90 (d, J = 19.3 Hz), 2.75 (d, J = 19.3 Hz), 2.17 (s), 2.15 (t, J = 7.4 Hz), 1.54 (m), 1.42 (s), 0.92 (t, J = 7.4 Hz); (+) ESIMS m/z 403.2 [M+H]+, calcd for C22H27O7 at m/z 403.1751.
Rugulosin A (6) – Yellow amorphous powder; 1H NMR (400 MHz, CD3OD): δ 7.50 (d, J = 1.0 Hz), 7.11 (s), 4.54 (d, J = 6.0 Hz), 3.48 (s), 2.84 (d, J = 6.0 Hz), 2.45 (s); (+) ESIMS m/z 543.4 [M+H]+, calcd for C30H23O10 at m/z 543.1286.
Comazaphilone C (7) – Yellow amorphous powder; 1H NMR (400 MHz, CDCl3): δ 6.54 (s), 6.48 (m), 5.91 (d, J = 16.0 Hz), 5.24 (s), 5.01 (d, J = 13.2 Hz), 4.87 (d, J = 13.2 Hz), 4.42 (t, J = 3.0 Hz), 3.79 (s), 2.71 (m), 2.26 (s), 1.85 (d, J = 7.0 Hz), 1.76 (s); (+) ESIMS m/z 417.3 [M+H]+, calcd for C22H25O8 at m/z 417.1544.
Questinol (8) – Yellow amorphous powder; 1H NMR (400 MHz, acetone-d6): δ 7.68 (d, J = 1.6 Hz), 7.36 (d, J = 2.3 Hz), 7.27 (d, J = 2.3 Hz), 6.94 (d, J = 1.6 Hz), 4.74 (s), 3.98 (s); (+) ESIMS m/z 301.1 [M+H]+, calcd for C16H13O6 at m/z 301.0707.
O-Coumaric acid (9) – White amorphous solid; 1H NMR (400 MHz, CD3OD): δ 7.92 (d, J = 16.0 Hz), 7.48 (dd, J = 8.1, 1.6 Hz), 7.19 (td, J = 7.8, 1.6 Hz), 6.84 (m), 6.82 (m), 6.53 (d, J = 16.0 Hz); (+) ESIMS m/z 165.2 [M+H]+, calcd for C9H9O3 at m/z 165.0546.
Asperflavin (10) – Amorphous soild; 1H NMR (400 MHz, CD3OD): δ 6.77 (s), 6.51 (d, J = 2.1 Hz), 6.42 (d, J = 2.1 Hz), 3.89 (s), 3.02 (d, J = 16.0 Hz), 2.96 (d, J = 16.0 Hz), 2.81 (d, J = 17.0 Hz), 2.71 (d, J = 17.0 Hz), 1.33 (s); (+) ESIMS m/z 289.3 [M+H]+, calcd for C16H17O5 at m/z 289.1071.
Flavomannin A (11) – Yellow amorphous powder; 1H NMR (400 MHz, CD3OD): δ 6.83 (s), 6.64 (s), 3.02 (d, J = 16.0 Hz), 2.98 (d, J = 16.0 Hz), 2.82 (d, J = 17.4 Hz), 2.73 (d, J = 17.4 Hz), 1.37 (s); (+) ESIMS m/z 547.4 [M+H]+, calcd for C30H27O10 at m/z 547.1599.
Results and Discussion
Chemical investigation of the culture extracts afforded the isolation of a new compound (1) together with known ones, 4-hydroxyemodin (2),4 kasanosin C (3),5 monomethyl-(S)-mitorubrin (4),6 comazaphilone A (5),7 rugulosin A (6),8 comazaphilone C (7),7 questinol (8),9 O-coumaric acid (9),10 asperflavin (10),11 and flavomannin A (11).12 Among them, kasanosin C (3), monomethyl-(S)-mitorubrin (4), comazaphilone A (5), and comazaphilone C (7) belong to azaphilone-type compounds and 4-hydroxyemodin (2), rugulosin A (6), questinol (8), asperflavin (10), and flavomannin A (11) are members of anthraquinone or anthrones.
compound 1 was isolated as a yellow amorphous powder. Its molecular formula was established to be C22H28O7 based on (+) HRESIMS, observed at m/z 404.1823. Its 1H-NMR spectrum showed olefinic protons signals at δH 6.10 and 6.04, an oxymethine at δH 4.74, a methoxy at δH 3.64, and four methyl signals at δH 2.22, 1.86, 1.80, and 0.93. In the 13C-NMR spectrum, two ketone carbons characteristic signals for azaphilones appeared at δC 206.0 and 196.2. In addition, signals for 2-methyl-4-hydroxy-6-methoxy benzoyl group, which is also present in kasanosin C (3), monomethyl-(S)-mitorubrin (4), and comazaphilone A (5), appeared in its 1H and 13C NMR. The complete chemical structure of 1 was established based on the interpretation of the 1H-1H COSY, HSQC, and HMBC data described subsequently. Two partial structures could be obtained through the 1H-1H COSY spectrum; CH(O)-CH2 at δH 4.74 and δH 2.86/2.58 corresponding to C-6/C-5 and CH2-CH2-CH3 corresponding to C-10/C-11/C-12. Strong HMBC correlations of the oxymethine proton H-6 with the ketone carbon C-8 at δC 196.2, the oxygenated quaternary carbon (C-7), the methyl carbon (C-9), and one of the double bond carbons (C-4a), and of a methyl group protons H-1 with C-8 and both double bond carbons suggested the presence of a 2,6-dimethyl-5-hydroxy-cyclohex-2-en-1-one moiety. Additionally, the methylene protons 2H-4 at δH 3.58 and 3.28 (each 1H, d, J = 16.6 Hz) showed strong HMBC correlations with another ketone carbon C-5 as well as C-4a,8a,10, indicating that 2-pentanone was attached to C-4a. In the previously proposed structure, the location of the 2-methyl-4-hydroxy-6-methoxy-benzoyl group could not be determined by HMBC correlations, expected due to the absence of protons around the connection site. Nonetheless, it was deduced to be attached to C-7 based on the NMR resonance and comparison with the analogues isolated from the same fungal strain and biosynthetic consideration. Accordingly, its planar structure was established as shown in the Fig. 1. Intriguingly, this compound is thought to be generated by the cleavage of the C1-C2 bond in the pyran ring of the typical azaphilone skeleton. To our knowledge, there is only one previous report on the isolation of the pyran-opened azaphilone, talarophilone, also isolated from Talaromyces sp.14 The key structural difference between talarophilone and 1 was the methoxylation at C-2' in the aromatic ring. (Fig. 2)
Acknowledgments
This work was supported by the National Research Foundation (NRF) of Korea (NRF-2021R1A2C1004958 and NRF-2022R1A4A3022401).
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
-
Jin, L.; Quan, C.; Hou, X.; Fan, S. Mar. Drugs 2016, 14, 76.
[https://doi.org/10.3390/md14040076]

-
Shin, H. J. Mar. Drugs 2020, 18, 230.
[https://doi.org/10.3390/md18050230]

-
Wang, Y.-T.; Xue, Y.-R.; Liu, C.-H. Mar. Drugs 2015, 13, 4594–4616.
[https://doi.org/10.3390/md13084594]

-
Morooka, N.; Nakano, S.; Itoi, N.; Ueno, Y. Agric. Biol. Chem. 1990, 54, 1247–1252.
[https://doi.org/10.1080/00021369.1990.10870097]

-
Li, L.-Q.; Yang, Y.-G.; Zeng, Y.; Zou, C.; Zhao, P.-J. Molecules 2010, 15, 3993–3997.
[https://doi.org/10.3390/molecules15063993]

-
Yamazaki, H.; Ōmura, S.; Tomoda, H. Chem. Pharm. Bull. 2010, 58, 829–832.
[https://doi.org/10.1248/cpb.58.829]

-
Gao, S.-S.; Li, X.-M.; Zhang, Y.; Li, C.-S.; Cui, C.-M.; Wang, B.-G. J. Nat. Prod. 2011, 74, 256–261.
[https://doi.org/10.1021/np100788h]

-
Song, F.; Dong, Y.; Wei, S.; Zhang, X.; Zhang, K.; Xu, X. Antibiotics 2022, 11, 222.
[https://doi.org/10.3390/antibiotics11020222]

-
Yang, X.; Kang, M.-C.; Li, Y.; Kim, E.-A.; Kang, S.-M.; Jeon, Y.-J. J. Microbiol. Biotechnol. 2014, 24, 1346–1353.
[https://doi.org/10.4014/jmb.1405.05035]

-
Kowczyk-Sadowy, M.; Świsłocka, R.; Lewandowska, H.; Piekut, J.; Lewandowski, W. Molecules 2015, 20, 3146–3169.
[https://doi.org/10.3390/molecules20023146]

-
Fujimoto, H.; Fujimaki, T.; Okuyama, E.; Yamazaki, M. Chem. Pharm. Bull. 1999, 47, 1426–1432.
[https://doi.org/10.1248/cpb.47.1426]

-
Bara, R.; Zerfass, I.; Aly, A. H.; Goldbach-Gecke, H.; Raghavan, V.; Sass, P.; Mandi, A.; Wray, V.; Polavarapu, P. L.; Pretsch, A.; Lin, W.; Kurtan, T.; Debbab, A.; Brotz-Oesterhelt, H.; Proksch, P. J. Med. Chem. 2013, 56, 3257−3272.
[https://doi.org/10.1021/jm301816a]

-
Xie, F.; Li, H.-T.; Chen, J.-Y.; Duan, H.-J.; Xia, D.-D.; Sun, Y.; Gao, Y.-H.; Zhou, H.; Ding, Z.-T. Tetrahedron Lett. 2022, 100, 153855.
[https://doi.org/10.1016/j.tetlet.2022.153855]