
Aster glehni Ethanol Extract Inhibits Inflammatory Responses Regulating Skin Barrier Molecules in Human Keratinocytes
Abstract
Prolonged skin inflammation is caused by disrupted skin barrier resulting in chronic inflammatory diseases such as atopic dermatitis. As a potent natural product with anti-inflammatory property, Aster glehni (A. glehni) is a traditional edible herb and has been used to treat diabetes or colitis-associated colon cancer. In present study, we figured out an additional effect of A. glehni ethanol extract (AGE) in pro-inflammatory cytokines-stimulated human keratinocytes. Mixture of tumor necrosis factor-alpha (TNF-α) and interferongamma (IFN-γ) was used to induce inflammatory responses in the HaCaT keratinocytes. AGE suppressed activation of ERK mitogen-activated protein kinase, nuclear factor (NF)-κB, and signal transducer and activator of transcription 1 and 3 (STAT1 and STAT3). The treatment of AGE inhibited mRNA expressions of proinflammatory cytokines in TNF-α and IFN-γ-stimulated HaCaT cells. Also, AGE induced up-regulated expressions of skin barrier molecules like filaggrin, loricrin, or ZO-1. We evaluated the effects of AGE on protein or mRNA expression levels using western blot or qRT-PCR, respectively. Taken together, these results suggest that the treatment of AGE exerts anti-inflammatory effect on keratinocytes through suppressing inflammatory signaling pathways and up-regulating skin molecules in HaCaT keratinocytes.
Keywords:
Aster glehni, HaCaT keratinocytes, Anti-inflammatory responses, Skin barrierIntroduction
Chronic skin inflammatory diseases are caused by an imbalance immune environment and easily induced by a disrupted skin barrier.1 As the organism’s first line of defense against the environment, skin is composed with two structures: epidermis and dermis.2 And, epidermis, outer layer of skin is consisted of different layers such as stratum basale, stratum spinosum, stratum granulosum, stratum lucidum, and stratum corneum.3 Keratinocytes are representative major cell type of the epidermis forming epidermal layers according to their differentiation stages.4 Since the interactions between keratinocytes and immune cells play a crucial role in regulating skin barrier homeostasis, immortalized human keratinocytes are a suitable model for studying skin inflammatory diseases in vitro.5 HaCaT cells were used for the investigating the effects of ethanol extract of A. glehni on skin inflammation which was induced by mixture of pro-inflammatory cytokines, mixture of tumor necrosis factor-alpha and interferongamma (TNF-α/IFN-γ).6
Aster glehni Franchet et Sckmidt, a conventional herb from Republic of Korea, is known for its a variety of bioactive constitutions and usages from edible ingredient to therapeutic agents.7 It has been used to treat excessive inflammation, obesity, nonalcoholic fatty liver, and colitis.8–10 Also, A. glehni methanol extract containing caffeoylquinic compounds showed protective effects on injured keratinocytes.11 In the present study, the authors established the objective of evaluating the effects of A. glehni 70% ethanol extract (AGE) on human keratinocytes.
Experimental
Chemicals and reagent – Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), penicillin, and streptomycin were got from Life Technologies Inc. (Grand Island, NY, USA). 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was bought from Sigma Chemical Co. (St. Louis, MO, USA). Dimethyl sulfoxide (DMSO) was purchased from Junsei Chemical Co. Ltd. (Tokyo, Japan).
Preparation of AGE – AGE was prepared as described.8,9 The dried A. glehni was refluxed with 70% ethanol for 6 h which was performed at 60℃. The extract was filtered using filter paper (Whatman qualitative filter paper, No. 4, 20-24 μm) and concentrated under decreased certain pressure. After the process, it was freeze-dried to get a solid extract powder.
Cell culture and sample treatment – One of keratinocytes cell-lines from human, HaCaT keratinocytes were cultured at 37℃ and 5% CO2 incubator in DMEM supplemented with 10% inactivated FBS, penicillin (100 U/mL), and streptomycin (100 μg/mL). Cells (1 × 105 cells/mL) were stimulated with mixture of TNF-α and IFN-γ (TNF-α/IFN-γ) which were used 10 ng/mL, respectively. The cells were shown the inflammatory responses by stimulation using TNF-α/IFN-γ for the specified time following the target markers.
Western blot analysis – Protein lysates were extracted from cell cultures (1 × 10⁵ cells/mL) using Pro-prep™ protein lysis buffer (Intron Biotechnology Inc., Gyeonggido, South Korea). Proteins were resolved using an 8–12% sodium dodecyl sulfate-polyacrylamide gel and then transferred onto polyvinylidene fluoride (PVDF) membranes following electrophoresis. Membranes were blocked for 30 minutes using 2.5–5% skim milk solution and subsequently incubated at 4°C overnight with primary antibodies diluted in Tris-buffered saline (TBS) containing 0.1% Tween-20. After removing the primary antibody by washing the membranes three times with TBS-T buffer, they were treated with horseradish peroxidase (HRP)-conjugated secondary antibodies (1:2500) at room temperature (25°C) for 2 hours. Following three additional washes in TBS-T, immunoreactive bands were visualized using enhanced chemiluminescence (ECL) solution (Absignal, Seoul, Republic of Korea) and captured on Xray film (Agfa, Belgium). Results represent the mean ± standard deviation (SD) of three separate experiments.
Real-Time Reverse Transcription Polymerase Chain Reaction (RT-PCR) – Total RNA was extracted from HaCaT keratinocyte cells using the Easy Blue RNA extraction kit in line with the manufacturer’s protocol. Complementary DNA (cDNA) was synthesized with cDNA reverse transcription kits (Life Technologies, Grand Island, NY, USA). The reverse transcription process was carried out using the GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA) and SYBR Premix Ex Taq. The synthesized cDNA fragments were approximately 200 base pairs in length. Amplification was performed using the StepOnePlus Real-Time PCR system (Applied Biosystems) with SYBR Green PCR Master Mix. Results were analyzed using the comparative threshold cycle (Ct) method and normalized to GAPDH optical density. SYBR Premix Ex Taq was obtained from Takara Bio (Shiga, Japan).
Statistical analysis – Data are expressed as the mean ± S.D. from repeated experiments which have been conducted three times. Significantly statistic differences were determined using ANOVA and Dunnett’s post hoc test, and p-values < 0.05 meant statistical significance.
Results and Discussion
Since keratinocytes make up about 90% of the epidermis, investigating of therapeutic candidates in the keratinocytes is necessary to figure out the effects or mechanisms of the ones.12 One of spontaneously immortalized human keratinocyte cell lines, HaCaT cells is proposed as a model for the study on skin inflammatory diseases like atopic dermatitis.13 Pro-inflammatory cytokines induce the inflammatory responses and impaired skin barriers mimicking features of atopic dermatitis as in vitro model.14,15 The authors found out the decreased expression levels of skin barrier molecules by mixture of proinflammatory cytokines (TNF-α/IFN-γ) in Fig. 1. Pretreatment of AGE significantly reversed both protein expression and mRNA expression of the skin barrier molecules including filaggrin, involucrin, and loricrin. The molecules are crucial epidermal barrier proteins in the cornified layer.16
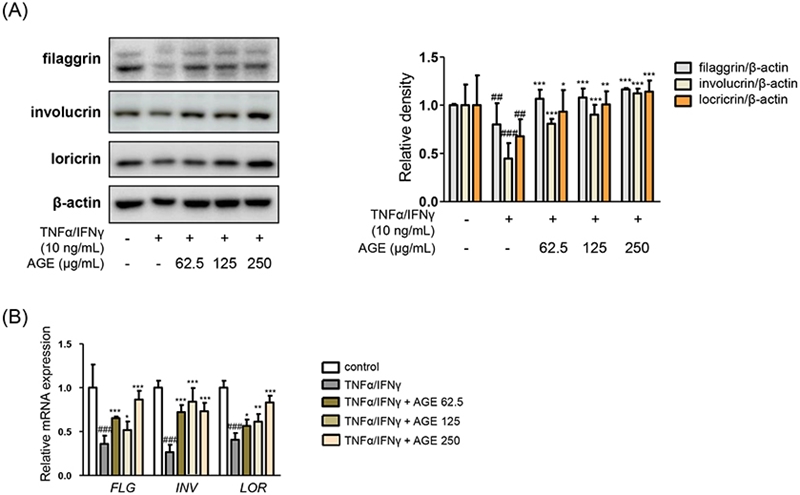
Effects of AGE on skin barrier molecules. HaCaT cells were treated with 62.5, 125, or 250 μg/mL of AGE for 1 h. And then the cells were incubated with mixture TNF-α/IFN-γ for 24 h. (A) The protein levels of filaggrin, involucrin, and loricrin were evaluated by immunoblot analysis with specific antibodies. (B) The mRNA expression levels of filaggrin, involucrin, and loricrin were analyzed by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Data were presented as the mean ± SD; *p < 0.05, **p < 0.01, ***p < 0.001 versus the TNF-α/IFN-γ-treated group, ##p < 0.01, ###p < 0.001 versus the control group.
Effects of AGE were shown in the different molecules consisting of tight junction. Pretreatment of AGE downregulated the protein expressions of matrix metalloproteinase (MMP) 1 and MMP9 in Fig. 2A. MMPs are zinc-containing endopeptidases which are able to degrade different components of extracellular matrix proteins.17 Though MMP1 and MMP9 are specifically classified into specifically collagenase and gelatinase, respectively, both of them are involved in integrity of skin barrier and inflammatory responses.18 Treatment of AGE down-regulated the expressions of MMPs as well as up-regulated the expressions of markers related with tight junction in Fig. 2 As members of composing of skin barrier structure, ZO-1, occludin, and claudin-1 were associated with adherens junctions.19 The mRNA expression levels of the components of tight junction were recovered by treatment of AGE comparing to the only TNF-α/IFN-γ-treated group (Fig. 2B).
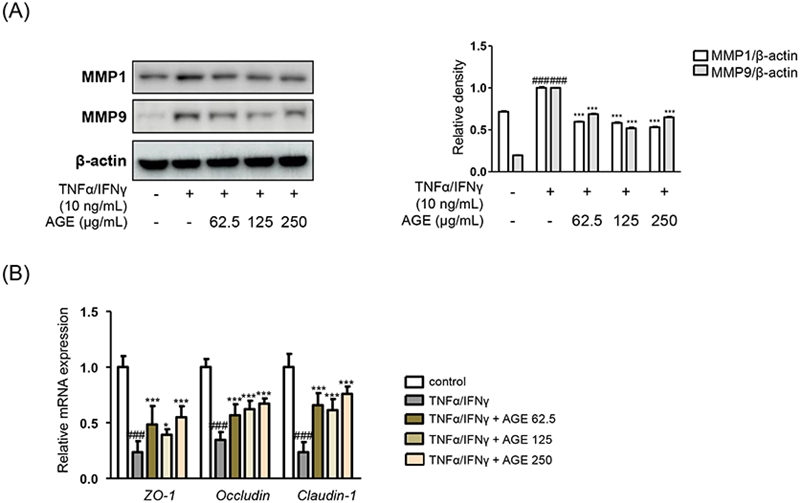
Effects of AGE on impaired skin barrier. (A) The protein levels of MMP1 and MMP9 were determined by immunoblot analysis with specific antibodies. (B) The mRNA expression levels of ZO-1, occludin, and claudin-1 were determined by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Data were figured out as the mean ± SD; *p < 0.05, ***p < 0.001 versus the TNF-α/ IFN-γ-treated group, ###p < 0.001 versus the control group.
Based on the results of AGE on regulating skin barrier with primary molecules, we evaluated the effects of AGE on inflammatory responses. It was examined on representative inflammatory signaling pathways, such as MAPK and NF-κB, as shown in Fig. 3. Since inflammatory skin state is accompanied with disruption of the barrier function, damaged barrier integrity is observed as a result of subsequent inflammatory responses.20 Effects of AGE in the protein expressions of extracellular signal-regulated kinase (ERK) 1/2 mitogen-activated protein kinase (MAPK) family were shown in Fig. 3. Also, results of AGE treatment in the protein expression of RAS/Raf/MAPK (MEK) 1/2 under the ERK cascade.21 Pretreatment of AGE suppressed the phosphorylation of MEK/ERK MAPK compared to the ones in only TNF-α/IFN-γ-treatment. Also, pretreatment of AGE inhibited the translocation of NF-κB p65, which is deeply involved in pro-inflammatory responses (Fig. 3B). As major regulator of epithelial homeostasis and inflammation, NF-κB activation results in increased cytokines production which led to pro-inflammatory responses.22 Increased mRNA expressions of pro-inflammatory cytokines caused by TNF-α/IFN-γ were suppressed by treatment of AGE: interleukin (IL)-1β, IL-6, IL-17, and TNF-α (Fig. 3D). Following the inhibitory effects of pro-inflammatory signaling pathways, effects of AGE were presented in the pro-inflammatory cytokines which led to skin inflammatory condition influencing epidermal morphologic characteristics and barrier function.23
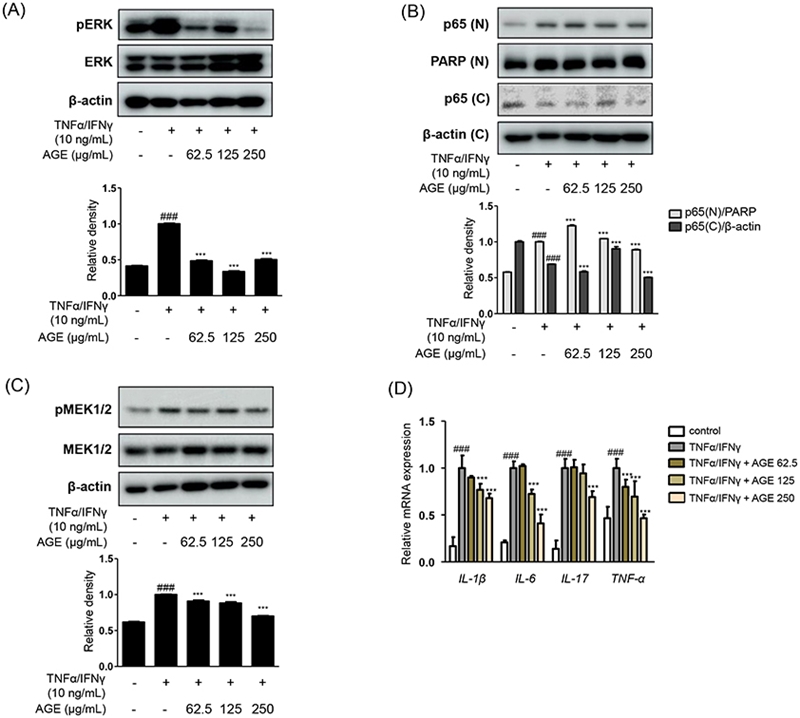
Effects of AGE on MEK-ERK MAPK and NF-κB pathways. The protein expressing levels of (A) ERK, (B) NF-κB p65, and (C) MEK1/2 phosphorylation were evaluated by immunoblot analysis with specific antibodies. (D) The mRNA expression levels of proinflammatory cytokines such as IL-1β, IL-6, IL-17, and TNF-α were determined by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Data were presented as the mean ± SD; ***p < 0.001 versus the TNF-α/IFN-γ-treated group, ###p < 0.001 versus the control group.
Effects of AGE treatment were investigated another signaling pathways: signal transducer and activator of transcription (STAT)s which are involved in skin inflammation.24 Phosphorylation of STAT1/STAT3 were significantly reduced at 250 μg/mL of AGE on each residues, serine and tyrosine (Fig. 4A and 4B).20,25 The recently emerging drugs for skin inflammatory diseases like atopic dermatitis have been focusing on inhibiting receptor signaling downstream the janus kinase (JAK)/STAT pathway.26 As new treatment option for patients who are suffered from moderate-to-severe atopic dermatitis, blocking IL-4/IL-13-signaling and JAK/STAT pathways such as Dupilumab,27 the effects of AGE were evaluated the phosphorylation of STAT as well as related regulator protein, suppressors of cytokine signaling (SOCS3).28 Along with the results shown in Fig. 3D, down-regulating effects of AGE were shown in protein expression of SOCS3 in Fig. 4C.
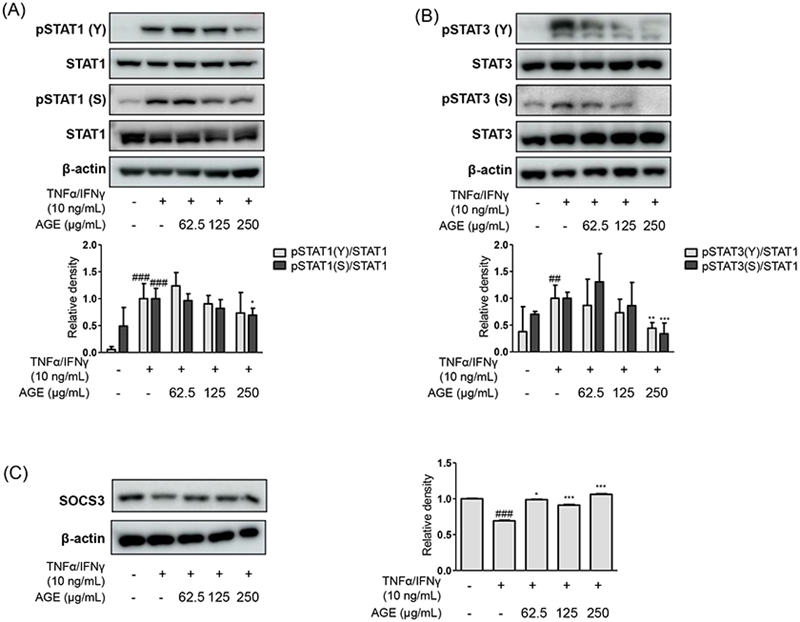
Effects of AGE on STAT1/3 signaling pathways. The protein levels of phosphorylation of (A) STAT1 and (B) STAT3 as well as (C) SOCS3 were examined under the certain immunoblot analysis using specific antibodies. Data were presented as the mean ± SD; *p < 0.05, **p < 0.01, ***p < 0.001 versus the TNF-α/IFN-γ-treated group, ##p < 0.01, ###p < 0.001 versus the control group.
Collecting the results, we evaluated the effects of AGE in down-regulating skin inflammatory responses as well as up-regulating skin barrier molecular in TNF-α/IFN-γ-stimulated HaCaT keratinocytes. We could suppose the effectiveness of AGE against inflammatory responses from potent candidate molecules of A. glehni ethanol extract: caffeoylquinic compounds, chlorogenic acid, luteolin, quercetin, saponins, or sesquiterpene lactones.8,29 These molecules showed anti-oxidant and anti-inflammatory effects inhibiting inflammatory cytokines and prostaglandins via pro-inflammatory signaling pathways such as MAPK and NF-κB.7,30 Furthermore, the effects of A. glehni methanol extract on regulating skin inflammation and strengthening the skin barrier11 enhance the potential of AGE as a suggestable and valuable agent against skin inflammation in vivo. Taken together, AGE could protect the hyper-inflammation in the skin by down-regulating pro-inflammatory pathway and up-regulating skin barrier molecules.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
-
Liu, Y.; Wang, H.; Taylor, M.; Cook, C.; Martínez-Berdeja, A.; North, J. P.; Harirchian, P.; Hailer, A. A.; Zhao, Z.; Ghadially, R.; Ricardo-Gonzalez, R. R.; Grekin, R. C.; Mauro, T. M.; Kim, E.; Choi, J.; Purdom, E.; Cho, R. J.; Cheng, J. B. Sci. immunol. 2022, 7, eabl9165.
[https://doi.org/10.1126/sciimmunol.abl9165]

-
Lee, S. H.; Jeong, S. K.; Ahn, S. K. Yonsei Med. J. 2006, 47, 293–306.
[https://doi.org/10.3349/ymj.2006.47.3.293]

-
Lefèvre-Utile, A.; Braun, C.; Haftek, M.; Aubin, F. Int. J. Mol. Sci. 2021, 22, 11676.
[https://doi.org/10.3390/ijms222111676]

-
Pondeljak, N.; Lugović-Mihić, L.; Tomic, L.; Parać, E.; Pedić, L.; Lazić-Mosler, E. Int. J. Mol. Sci. 2023, 25, 236.
[https://doi.org/10.3390/ijms25010236]

-
Gallegos-Alcalá, P.; Jiménez, M.; Cervantes-García, D.; Salinas, E. Int. J. Mol. Sci. 2021, 22, 10661.
[https://doi.org/10.3390/ijms221910661]

-
Yang, J.-H.; Yoo, J.-M.; Lee, E.; Lee, B.; Cho, W.-K.; Park, K.-I.; Yeul Ma, J. Y. J. Ethnopharmacol. 2018, 211, 217–223.
[https://doi.org/10.1016/j.jep.2017.09.041]

-
Nugroho, A.; Kim, M.-H.; Choi, J.; Choi, J. S.; Jung, W. T.; Lee, K.-T.; Park, H.-J. Arch. Pharm. Res. 2012, 35, 423–430.
[https://doi.org/10.1007/s12272-012-0304-7]

-
Lee, H.-M.; Yang, G.; Ahn, T.-G.; Kim, M.-D.; Nugroho, A.; Park, H.-J.; Lee, K.-T.; Park, W.; An, H.-J. Evid. Based Complement. Alternat. Med. 2013, 2013, 859624.
[https://doi.org/10.1155/2013/859624]

-
Jin, B.-R.; Chung, K.-S.; Lee, M.; An, H.-J. Biology 2020, 9, 24.
[https://doi.org/10.3390/biology9020024]

-
Lee, Y.-J.; Jang, Y.-N.; Han, Y.-M.; Kim, H.-M.; Jeong, J.-M.; Son, M. J.; Jin, C. B.; Kim, H. J.; Seo, H. S. PPAR Res. 2017, 2017, 3912567.
[https://doi.org/10.1155/2017/3912567]

- Lee, Y.-J.; Jang, Y.-N.; Han, Y.-M.; Kim, H.-M.; Jin, C.; Kim, H. J.; Seo, H. S. Evid. Based Complement. Alternat. Med. 2018, 2018, 9616574.
-
McGrath, J. A.; Eady, R.; Pope, F. Rook’s textbook of dermatology - Anatomy and organization of human skin, Blackwell Pub; United States, 2004, pp 2–80.
[https://doi.org/10.1002/9780470750520.ch3]

-
Colombo, I.; Sangiovanni, E.; Maggio, R.; Mattozzi, C.; Zava, S.; Corbett, Y.; Fumagalli, M.; Carlino, C.; Corsetto, P. A.; Scaccabarozzi, D.; Calvieri, S.; Gismondi, A.; Taramelli, D.; Dell'Agli, M. Mediators Inflamm. 2017, 2017, 7435621.
[https://doi.org/10.1155/2017/7435621]

-
De Vuyst, E.; Salmon, M.; Evrard, C.; Lambert de Rouvroit, C.; Poumay, Y. Front. Med. 2017, 4, 119.
[https://doi.org/10.3389/fmed.2017.00119]

-
Yoshida, T.; Beck, L. A.; De Benedetto, A. Allergol. Int. 2022, 71, 3–13.
[https://doi.org/10.1016/j.alit.2021.11.006]

-
Candi, E.; Schmidt, R.; Melino, G. Nat. Rev. Mol. Cell Biol 2005, 6, 328–340.
[https://doi.org/10.1038/nrm1619]

-
Pittayapruek, P.; Meephansan, J.; Prapapan, O.; Komine, M.; Ohtsuki, M. Int. J. Mol. Sci. 2016, 17, 868.
[https://doi.org/10.3390/ijms17060868]

-
Philips, N.; Auler, S.; Hugo, R.; Gonzalez, S. Enzyme Res. 2011, 2011, 427285.
[https://doi.org/10.4061/2011/427285]

-
Pummi, K.; Malminen, M.; Aho, H.; Karvonen, S. L.; Peltonen, J.; Peltonen, S. J. Invest. Dermatol. 2001, 117, 1050–1058.
[https://doi.org/10.1046/j.0022-202x.2001.01493.x]

-
Baker, P.; Huang, C.; Radi, R.; Moll, S. B.; Jules, E.; Arbiser, J. L. Cells 2023, 12, 2745.
[https://doi.org/10.3390/cells12232745]

- Guo, Y.-J.; Pan, W. -W.; Liu, S.-B.; Shen, Z.-F.; Xu, Y.; Hu, L.-L. Exp. Ther. Med. 2020, 19, 1997–2007.
-
Wullaert, A.; Bonnet, M. C.; Pasparakis, M. Cell Res. 2011, 21, 146–158.
[https://doi.org/10.1038/cr.2010.175]

-
Danso, M. O.; van Drongelen, V.; Mulder, A.; van Esch, J.; Scott, H.; van Smeden, J.; El Ghalbzouri, A.; Bouwstra, J. A. J. Invest. Dermatol. 2014, 134, 1941–1950.
[https://doi.org/10.1038/jid.2014.83]

-
Villarino, A. V.; Kanno, Y.; O'Shea, J. J. Nat. Immunol. 2017, 18, 374–384.
[https://doi.org/10.1038/ni.3691]

-
Archer, N.; Lee, S. K.; Ortines, R. V.; Wang, Y.; Liu, H.; Miller, R. J.; Dillen, C. A.; Marchitto, M.; Ashbaugh, A. G.; Uppal, A. J. Immunol. 2017, 198, 197–204.
[https://doi.org/10.4049/jimmunol.198.Supp.197.4]

-
Nakashima, C.; Yanagihara, S.; Otsuka, A. Allergol. Int. 2022, 71, 40–46.
[https://doi.org/10.1016/j.alit.2021.10.004]

-
Seegraber, M.; Srour, J.; Walter, A.; Knop, M.; Wollenberg, A. Expert Rev. Clin. Pharmacol. 2018, 11, 467–474.
[https://doi.org/10.1080/17512433.2018.1449642]

-
Croker, B. A.; Kiu, H.; Nicholson, S. E. Semin. Cell Dev. Biol. 2008, 19, 414–422.
[https://doi.org/10.1016/j.semcdb.2008.07.010]

-
Naveed, M.; Hejazi, V.; Abbas, M.; Kamboh, A. A.; Khan, G. J.; Shumzaid, M.; Ahmad, F.; Babazadeh, D.; FangFang, X.; Modarresi-Ghazani, F.; WenHua, L.; XiaoHui, Z. Biomed. Pharmacother. 2018, 97, 67–74.
[https://doi.org/10.1016/j.biopha.2017.10.064]

-
Jeong, J.; Lim, M. K.; Han, E. H.; Lee, S.-H.; Kang, S.; Lee, S. Food Sci. Biotechnol. 2022, 31, 1729–1739.
[https://doi.org/10.1007/s10068-022-01153-5]
