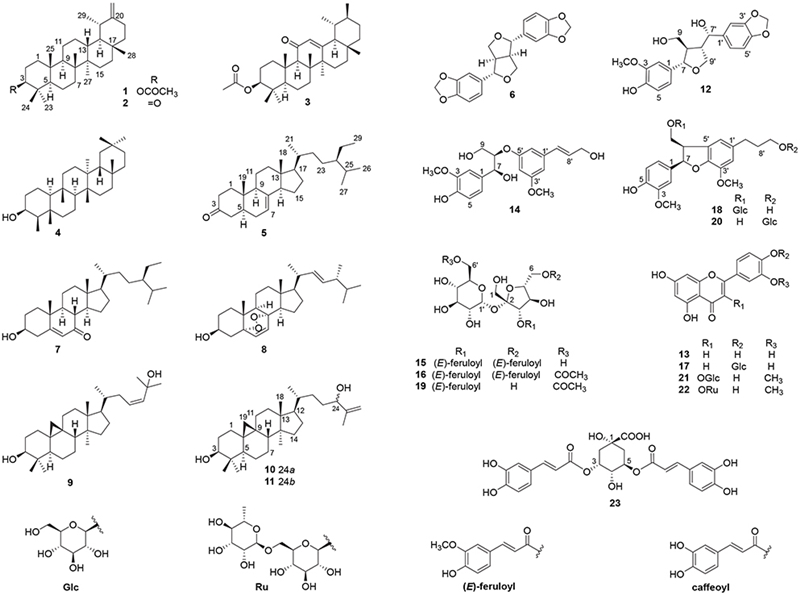
Anti-inflammatory and Cytotoxic Effects of Compounds from the Aerial Parts of Achillea alpina L.
Abstract
Achillea alpina L. is a medicinal herb belonging to the Asteraceae family. Its leaves are used as a vegetable and for treating stomach ailments in Korea. Our current research focused on isolating and identifying the chemical constituents from the aerial parts of this plant, leading to the discovery of 23 compounds, including 4 pentacyclic triterpenes (1‒4), 3 steroids (5, 7, and 8), 3 cycloartanes (9‒11), 5 lignans (6, 12, 14, 20, and 18), 4 flavonoids (13, 17, 21, and 22), 3 feruloyl sucroses (16, 18, and 19), and 1 caffeoyl quinic acid (23). The structures of these compounds were determined through spectral data analysis (1D and 2D NMR spectroscopy) and comparison with relevant literature. Notably, the isolated sterols (5, 7, and 8) and flavone (13) inhibited NO production in LPS-stimulated RAW 264.7 cells with IC50 values ranging from 8.69 to 43.17 μM. Additionally, sterol (5) exhibited cytotoxic effects on HeLa and HL-60 (human leukemia) cancer cell lines with ED50 values ranging from 53.57 to 78.33 μM.
Keywords:
Achillea alpina L., Asteraceae, Anti-inflammation, CytotoxicityIntroduction
The genus Achillea (the family Asteraceae) comprises over 130 species from subtropical to tropical regions, thriving in various terrains.1 Several species in this genus have a long history of use in traditional medicine across many countries. For example, A. millefolium L. is employed to treat appetite loss, skin diseases, minor wounds, urinary tract issues, and gastrointestinal problems.1,2
Achillea alpina L. is abundant in Korea, China, Japan, Russia, and Mongolia.3 In Korean folk medicine, its young leaves are used as vegetables and to treat stomach ailments.4 Additionally, Traditional Chinese medicine uses this plant for pain relief and to promote blood circulation.5 Previous phytochemical studies have reported various metabolites from this plant, including terpenes,5–7 lignans,8,9 flavonoids,9 alkaloids,10 and polyacetylenes.8,9 The sesquiterpene lactones isolated from A. alpina exhibited antidiabetic activity by enhancing glucose consumption and reducing insulin resistance in palmitic acid-treated HepG2 cells.5,6 Meanwhile, its phenolic compounds showed anti-melanogenic and anti-oxidant effects, thereby demonstrating anti-aging and skin-whitening properties.4 In addition, alkamides isolated from this plant have demonstrated neuroprotective activity on 6-hydroxydopamine (6-OHDA)-induced cell death in human neuroblastoma SH-SY5Y cells with ED50 values of 3.16–24.75 μM.10 To further elucidate the chemical richness and pharmacological effects of this plant, our current research aimed to isolate and characterize the secondary metabolites from the aerial parts of A. alpina. We then evaluated their NO production inhibitory activity in LPS-stimulated RAW 264.7 cells and their cytotoxicity against HeLa and HL-60 cancer cell lines.
Experimental
General experiment procedures – Specific optical rotation was determined using a JASCO P-2000 digital polarimeter (JASCO Corporation, Tokyo, Japan). UV spectra were recorded with a Thermo spectrophotometer, while IR spectra were obtained using a JASCO FT/IR-4100 spectrometer. The 1D and 2D NMR spectra were acquired on Varian Unity Inova 400 MHz (Varian, Inc., California, USA) and Bruker Ascend™ 500 MHz (Bruker Corporation, Massachusetts, USA) spectrometers, using tetramethylsilane (TMS) as an internal standard; chemical shifts are expressed in δ values (ppm). High-performance liquid chromatography was executed on a Waters 2487 controller system with a UV detector (UV/VIS-156). Silica gel (Merck, 0.040-0.063 mm), RPC-18 (Merck, 17 mesh), and Sephadex LH-20 (Pharmacia Company) were utilized for column chromatography (CC). For thin-layer chromatography (TLC), RPC-18 F254s and silica gel F254 (Merck) plates were employed, with compounds visualized by spraying with 10% H2SO4 and heating for 5 minutes.
Chemical and reagents – DMEM, RPMI, and DPBS originated from Thermo Fisher Scientific (Gibco Laboratories, Grand Island, NY, U.S.A). FBS was the brand of Biotechnics Research Inc. (Lake Forest, CA, U.S.A). LPS, MTT, penicillin, and doxorubicin hydrochloride belong to Sigma-Aldrich (Merck KGaA, St. Louis, Missouri, USA).
Plant materials – The aerial parts of A. alpina were collected from the medicinal garden at the College of Pharmacy, Daegu Catholic University, Republic of Korea in July 2021. The plant materials were identified and preserved voucher specimens (CUD-3001) by Professor Byung Sun Min at the Herbarium of College of Pharmacy at Daegu Catholic University, Republic of Korea.
Extraction and isolation – The aerial parts of A. alpina (17.7 kg) were extracted with methanol (MeOH) (20 L × 3 times). The MeOH extract (2.1 kg) was obtained by removing the MeOH under vacuum. This extract was then suspended in distilled water and sequentially partitioned using solvents of increasing polarity, including n-hexane, CH2Cl2, and EtOAc, to obtain the n-hexane (260.6 g), the CH2Cl2 (396.8 g), and the EtOAc fractions (41.6 g), respectively, along with the aqueous phase.
The CH2Cl2 fraction (396.8 g) was preliminarily separated using silica gel column chromatography (CC) with n-hexane: acetone (stepwise, 100:1 → 100% acetone) to produce 10 fractions M1‒M10. Fractions M3 (6.1 g) and M5 (2.5 g) were further separated into smaller fractions (M3.1‒M3.6) and (M5.1‒M5.6), respectively, using silica gel CC (n-hexane:acetone, 30:1). Compound 1 (17.0 mg) was recrystallized from M3.3 (864.2 mg) in n-hexane. Compound 2 (8.0 mg) was obtained from M3.4 (500.1 mg) using a silica gel column (n-hexane:acetone, 32:1). Using silica gel CC with the mobile phase of n-hexane:acetone mixture but varying the solvent system ratio, specifically 28:1 (n-hexane:acetone) for M5.1 (67.2 mg), 25:1 for M5.3 (124.3 mg), and 20:1 for M5.5 (256.9 mg), led to the isolation of compounds 3 (1.0 mg), 4 (3.9 mg), and 5 (12.6 mg), respectively. M6 (12.4 g) was also divided into 10 sub-fractions (M6.1‒M6.10) using a silica gel column eluted with n-hexane:acetone (50:1). Compound 6 (68.2 mg) was crystallized from sub-fraction M6.2 (2.2 g) in n-hexane. M8 (9.1 g) was fractionated using silica gel CC (CH2Cl2:acetone, 50:1) to obtain 12 sub-fractions (M8.1‒ M8.12). M8.1 (534.5 mg) and M8.3 (1.2 g) were separated using normal-phase CC (n-hexane:acetone, 15:1), yielding compounds 7 (8.6 mg) and 8 (12.5 mg), respectively. M8.9 (1.6 g) was purified using silica gel CC (CH2Cl2:acetone, 40:1) to obtain 9 (10.2 mg). M8.10 (42.3 mg) was a mixture of two isomers, 10 (6.0 mg) and 11 (12.0 mg), which were subsequently separated using a long normal-phase column with CH2Cl2:acetone (30:1) as the mobile phase.
The EtOAc fraction (41.6 g) underwent silica gel CC, with elution sequentially using CH2Cl2:MeOH (20:1 → 100% MeOH), resulting in 14 distinct fractions (E1–E14). Among these, E4 (1.6 g) was further purified via silica gel CC, using CH2Cl2:MeOH (22:1) as the elution system, to yield sub-fractions E4.1‒E4.9. From E4.4 (150.2 mg), separation was carried out using RPC-18 CC with a mobile phase of MeOH:H2O (1:1), successfully isolating compound 12 (2.1 mg). Fraction E5 (3.2 g) was divided into 6 sub-fractions (E5.1‒E5.6) using a normal-phase column eluted with CH2Cl2:MeOH (15:1). E5.5 (1.2 g) was recrystallized using CH2Cl2, resulting in two portions: E5.5.1 (crystalline) and E5.5.2 (non-crystalline). Subfraction 5.5.1 (113.5 mg) was further processed by repeated washing with CH2Cl2:MeOH (1:1), yielding purified compound 13 (62.6 mg). Sub-fraction E5.5.2 (950.3 mg) underwent RPC-18 CC (MeOH:H2O, 1:4), followed by Sephadex CC using (MeOH:H2O, 1:1) to isolate compound 14 (4.8 mg). Additionally, fractions E8 (6.3 g) and E10 (5.2 g) were divided into five (E8.1‒E8.5) and six subfractions (E10.1‒E10.6), respectively, using silica gel CC with CH2Cl2:MeOH (6:1) as the eluent. Sub-fractions E8.1 (1.1 g) and E8.3 (1.2 g) were chromatographed by RPC-18 CC (MeOH:H2O,1:4), resulting in the separation of sub-fractions E8.1.1‒8.1.6 and E8.3.1‒8.3.5. E8.3.1 (181.2 mg) and E8.3.5 (56.3 mg) were further purified by HPLC using 50% MeOH in water, yielding compounds 15 (6.0 mg, tR = 21.5 minutes) and 16 (5.0 mg, tR = 26.8 minutes), respectively. Sub-fraction E10.1 (1.6 g) was then separated by RPC-18 CC (MeOH:H2O, stepwise, 1:4 → 1:1) to yield compound 17 (7.0 mg) and sub-fractions E10.1.1‒E10.1.3. Using Sephadex CC (MeOH:H2O, 1:1), compound 18 (9.8 mg) was obtained from E10.1.2 (289.1 mg) and compound 19 (3.1 mg) from E10.1.3 (92.9 mg). Fractions E11 (2.3 g), E12 (2.4 g), and E14 (3.1 g) were fractionated by silica gel CC using CH2Cl2:MeOH (5:1) to yield six (E11.1‒E11.6), four (E12.1‒E12.3), and two sub-fractions (E14.1‒E14.2), respectively. Compounds 20 (4.9 mg) and 21 (30.0 mg) were separated from subfraction E11.2 (900.2 mg), compound 22 (9.6 mg) was obtained from E12.2.2 (421.7 mg), and compound 23 (15.6 mg) was obtained from E14.1 (534.8 mg) using RPC-18 CC (MeOH:H2O, 1:2).
Taraxasteryl acetate (1) – Colorless needle; 1H-NMR (400 MHz, CDCl3): δ 4.61 (1H, s, H-30b), 4.60 (1H, s, H-30a), 4.48 (1H, dd, J = 10.0, 6.3 Hz, H-3), 2.04 (3H, s, COCH3), 1.02 (6H, s, CH3-25, CH3-23), 0.93 (3H, s, CH3-27), 0.87 (3H, s, CH3-26), 0.84 (3H, s, CH3-24), 0.83 (6H, s, CH3-29, CH3-28); 13C-NMR (100 MHz, CDCl3) δ (ppm): 170.8 (COCH3), 154.5 (C-20), 106.9 (C-30), 80.8 (C-3), 55.3 (C-5), 50.2 (C-9), 48.5 (C-18), 41.9 (C-14), 40.8 (C-8), 39.2 (C-22), 39.0 (C-16), 38.7 (C-13), 38.2 (C-1), 38.1 (C-19), 37.6 (C-4), 36.9 (C-10), 34.4 (C-17), 33.8 (C-7), 27.8 (C-23), 26.5 (C-15), 26.0 (C-28), 25.5 (C-12), 25.3 (C-21), 23.5 (C-2), 21.3 (C-11), 21.2 (CH3CO), 19.3 (C-29), 18.0 (C-6), 16.3 (C-24), 16.2 (C-26), 15.7 (C-25), 14.6 (C-27).11
Taraxast-20(30)-en-3-one (2) – Colorless needle; 1HNMR (400 MHz, CDCl3): δ 4.62 (1H, s, H-30b), 4.60 (1H, s, H-30a), 1.07 (3H, s, CH3-24), 1.05 (3H, s, CH3-23), 1.02 (6H, m, CH3-28, CH3-29), 0.95 (3H, s, CH3-26), 0.94 (3H, s, CH3-27), 0.86 (3H, s, CH3-25); 13C-NMR (100 MHz, CDCl3): δ 218.3 (C-3), 154.7 (C-20), 107.4 (C-30), 55.1 (C-5), 50.0 (C-9), 48.8 (C-18), 47.5 (C-4), 42.3 (C-14), 41.0 (C-8), 39.8 (C-1), 39.5 (C-13), 39.4 (C-19), 39.0 (C-22), 38.4 (C-16), 37.0 (C-10), 34.7 (C-17), 34.3 (C-2), 33.5 (C-7), 26.9 (C-15), 26.8 (C-12), 26.3 (C-29), 25.8 (C-21), 25.6 (C-23), 22.1 (C-24), 21.2 (C-11), 19.8 (C-6), 19.7 (C-28), 16.3 (C-26), 15.9 (C-25), 14.8 (C-27).12
Neoilexonol acetate (3) – Colorless needle; 1H-NMR (400 MHz, CDCl3): δ 5.54 (1H, s, H-12), 4.52 (1H, dd, J = 11.9, 4.8 Hz, H-3), 2.05 (3H, s, COCH3), 1.54 (3H, s, CH3-27), 1.29 (3H, s, CH3-26), 1.24 (3H, s, CH3-25), 1.19 (3H, s, CH3-23), 1.16 (3H, s, CH3-24), 0.87 (6H, m, H-28, H-30), 0.81 (3H, d, J = 6.9 Hz, H-29); 13C-NMR (100 MHz, CDCl3): δ 199.7 (C-11), 171.0 (COCH3), 165.0 (C-13), 130.4 (C-12), 80.7 (C-13), 61.5 (C-9), 59.1 (C-18), 55.1 (C-5), 45.2 (C-17), 43.7 (C-14), 41.0 (C-8), 39.4 (C-20), 39.3 (C-19), 38.9 (C-1), 38.1 (C-4), 36.9 (C-10), 34.0 (C-21), 32.9 (C-7), 30.9 (C-22), 28.9 (C-15), 28.1 (C-23), 28.1 (C-28), 27.6 (C-27), 27.3 (C-16), 23.6 (C-2), 21.4 (C-28), 21.2 (COCH3), 20.6 (C-26), 18.6 (C-6), 17.5 (C-29), 16.8 (C-24), 16.6 (C-25).13
Epifriedelanol (4) – Colorless needle; 1H-NMR (400 MHz, CDCl3): δ 3.73 (1H, br s, H-3), 1.17 (3H, s, CH3-30), 1.01 (3H, s, CH3-26), 0.99 (3H, s, CH3-28), 0.98 (3H, s, CH3-27), 0.96 (3H, s, CH3-24), 0.94 (6H, m, CH3-23, CH3-29), 0.86 (3H, s, CH3-25); 13C-NMR (100 MHz, CDCl3): δ 72.7 (C-3), 61.3 (C-10), 53.1 (C-8), 49.1 (C-4), 42.7 (C-18), 41.7 (C-6), 39.6 (C-13), 39.2 (C-5), 38.3 (C-9), 37.8 (C-13), 37.1 (C-5), 36.0 (C-16), 35.5 (C-19), 35.3 (C-11), 35.1 (C-2), 35.0 (C-29), 32.8 (C-21), 32.3 (C-15), 32.0 (C-30), 31.7 (C-28), 30.6 (C-12), 30.0 (C-17), 28.1 (C-20), 20.1 (C-27), 18.6 (C-26), 18.2 (C-25), 17.5 (C-7), 16.4 (C-24), 15.7 (C-1), 11.6 (C-23).14
Stigmast-7-en-3-one (5) – Colorless oil; 1H-NMR (400 MHz, CDCl3): δ 5.17 (1H, s, H-7), 1.00 (3H, s, CH3-19), 0.92 (3H, d, J = 6.4 Hz, CH3-21), 0.82 (9H, m, CH3-27, CH3-28, CH3-29), 0.55 (3H, s, CH3-18); 13C-NMR (100 MHz, CDCl3): δ 212.1 (C-3), 139.7 (C-8), 117.1 (C-7), 56.2 (C-17), 55.1 (C-14), 49.0 (C-5), 46.0 (C-9), 44.4 (C-24), 43.5 (C-13), 43.0 (C-4), 39.6 (C-10), 38.9 (C-12), 38.3 (C-1), 36.7 (C-20), 34.6 (C-20), 34.0 (C-22), 30.2 (C-25), 29.3 (C-6), 28.1 (C-16), 26.4 (C-15), 23.2 (C-23), 23.1 (C-28), 21.9 (C-11), 20.0 (C-26), 19.2 (C-27), 19.1 (C-21), 12.6 (C-19), 12.1 (C-29), 12.1 (C-18).15
Sesamin (6) – Colorless needle; 1H-NMR (400 MHz, CDCl3): δ 6.84 (2H, s, H-2, H-2′), 6.79 (4H, m, H-5, H-6, H-5′, H-6′), 5.95 (4H, s, OCH2O x 2), 4.71 (2H, m, H-7, H-7′), 4.23 (2H, m, H-9a, H-9′a), 3.86 (2H, m, H-9b, H-9′b), 3.05 (2H, m, H-8, H-8′); 13C-NMR (100 MHz, CDCl3): δ 148.1 (C-3, C-3′), 147.2 (C-4, C-4′), 135.2 (C-1, C-1′), 119.5 (C-6, C-6′), 108.3 (C-5, C-5′), 106.2 (C-2, C-2′), 101.2 (OCH2O x 2), 85.9 (C-7, C-7′), 71.9 (C-9, C-9′), 54.5 (C-8, C-8′).16
7-Keto-β-sitosterol (7) – Colorless oil; 1H-NMR (400 MHz, CDCl3): δ 5.65 (1H, s, H-6), 3.61 (1H, m, H-3), 1.16 (3H, s, CH3-19), 0.89 (3H, d, J = 6.9 Hz, CH3-21), 0.81 (3H, t, J = 7.3 Hz, CH3-29), 0.79 (3H, d, J = 6.9 Hz, CH3-26), 0.77 (3H, d, J = 6.9 Hz, CH3-27), 0.64 (3H, s, CH3-18); 13C-NMR (100 MHz, CDCl3): δ 202.3 (C-7), 165.4 (C-5), 125.8 (C-6), 70.2 (C-3), 54.5 (C-17), 49.8 (C-14), 49.7 (C-9), 45.6 (C-24), 45.2 (C-8), 42.9 (C-13), 41.6 (C-4), 38.5 (C-12), 38.1 (C-10), 36.2 (C-1), 35.9 (C-20), 33.7 (C-22), 30.9 (C-2), 28.9 (C-25), 28.4 (C-16), 26.1 (C-15), 25.9 (C-23), 22.9 (C-28), 21.0 (C-11), 19.6 (C-27), 18.9 (C-26), 18.7 (C-21), 17.1 (C-19), 11.8 (C-29), 11.8 (C-18).17
Ergosterol peroxide (8) – Colorless oil; 1H-NMR (400 MHz, CDCl3): δ 6.48 (1H, d, J = 8.5 Hz, H-7), 6.22 (1H, d, J = 8.5 Hz, H-6), 5.20 (1H, dd, J = 15.2, 7.4 Hz, H-22), 5.12 (1H, dd, J = 15.2, 8.2 Hz, H-23), 3.93 (1H, m, H-3), 0.98 (3H, d, J = 6.6 Hz, CH3-21), 0.89 (3H, J = 6.8 Hz, CH3-28), 0.86 (3H, s, CH3-19), 0.80 (3H, s, CH3-18), 0.79 (6H, d, J = 3.2 Hz, CH3-26, 27); 13C-NMR (100 MHz, CDCl3): δ 135.3 (C-6), 135.1 (C-22), 132.1 (C-23), 130.6 (C-7), 82.1 (C-5), 79.3 (C-8), 66.2 (C-3), 56.0 (C-17), 51.5 (C-14), 50.9 (C-9), 44.4 (C-13), 42.6 (C-24), 39.6 (C-20), 39.2 (C-12), 36.8 (C-20), 36.7 (C-4), 34.6 (C-10), 32.9 (C-1), 29.9 (C-25), 28.5 (C-2), 23.3 (C-16), 20.7 (C-15), 20.5 (C-21), 19.8 (C-11), 19.5 (C-27), 18.0 (C-19), 17.4 (C-28), 12.7 (C-18).18
9,19-Cyclolanost-23-ene-3β,24-diol (9) –Colorless needle; 1H-NMR (400 MHz, CDCl3): δ 5.58 (2H, m, H-23, H-24), 3.26 (1H, dd, J = 10.3, 4.2 Hz, H-3), 1.30 (6H, s, CH3-26, CH3-27), 0.95 (6H, s, CH3-18, CH3-29), 0.87 (3H, s, CH3-28), 0.84 (3H, d, J = 6.4 Hz, CH3-21), 0.79 (3H, s, CH3-30), 0.54 (1H, d, J = 3.8 Hz, H-19a), 0.31 (1H, d, J = 3.8 Hz, H-19b); 13C-NMR (100 MHz, CDCl3): δ 139.8 (C-24), 126.1 (C-23), 79.3 (C-3), 71.2 (C-25), 52.5 (C-17), 49.3 (C-14), 48.4 (C-8), 47.6 (C-5), 45.8 (C-13), 50.0 (C-4), 39.5 (C-22), 36.9 (C-20), 36.1 (C-15), 33.3 (C-12), 32.4 (C-1), 30.9 (C-2), 30.5 (C-26), 30.4 (C-19, 27), 28.5 (C-16), 26.9 (C-11), 26.6 (C-7), 26.5 (C-10), 25.9 (C-29), 21.6 (C-6), 20.5 (C-9), 19.8 (C-28), 18.8 (C-21), 18.5 (C-18), 14.5 (C-30).19
24α-9,19-Cyclolanost-25-ene-3β,24-diol (10) – Colorless needle; 1H-NMR (400 MHz, CDCl3): δ 4.91 (1H, s, H-26a), 4.83 (1H, s, H-26b), 4.01 (1H, t, J = 6.1 Hz, H-24), 3.27 (1H, m, H-3), 1.72 (3H, s, CH3-27), 0.96 (6H, s, H-28, 30), 0.87 (3H, s, CH3-18), 0.86 (3H, d, J = 6.9 Hz, CH3-21), 0.80 (3H, s, CH3-29), 0.55 (1H, d, J = 4.1 Hz, H-19a), 0.32 (1H, d, J = 4.1 Hz, H-19b); 13C-NMR (100 MHz, CDCl3): δ 147.6 (C-25), 111.5 (C-26), 80.0 (C-3), 76.9 (C-24), 52.3 (C-17), 48.9 (C-14), 48.1 (C-8), 47.3 (C-5), 45.4 (C-13), 40.6 (C-4), 36.1 (C-20), 35.7 (C-15), 33.0 (C-12), 32.1 (C-22), 32.0 (C-1), 31.7 (C-23), 30.5 (C-2), 30.0 (C-19), 28.2 (C-16), 26.6 (C-11), 26.3 (C-10), 26.2 (C-7), 25.6 (C-29), 21.3 (C-6), 20.1 (C-9), 19.5 (C-28), 18.5 (C-21), 18.2 (C-18), 17.4 (C-27), 14.2 (C-30).19
24β-9,19-Cyclolanost-25-ene-3β,24-diol (11) – Colorless needle; 1H-NMR (400 MHz, CDCl3): δ 4.93 (1H, s, Ha-26), 4.83 (1H, s, Hb-26), 4.02 (1H, m, H-24), 3.28 (1H, m, H-3), 1.73 (3H, s, H-27), 0.98 (6H, s, CH3-28, CH3-30), 0.88 (3H, s, CH3-18), 0.87 (3H, d, J = 6.8 Hz, CH3-21), 0.80 (3H, s, CH3-29), 0.55 (1H, d, J = 3.9 Hz, H-19a), 0.33 (1H, d, J = 3.9 Hz, H-19b); 13C-NMR (100 MHz, CDCl3): δ 147.9 (C-25), 111.0 (C-26), 80.0 (C-3), 76.5 (C-24), 52.3 (C-17), 49.0 (C-14), 48.1 (C-8), 47.3 (C-5), 45.4 (C-13), 40.6 (C-4), 36.1 (C-20), 35.7 (C-15), 33.0 (C-12), 32.1 (C-22), 32.1 (C-1), 31.8 (C-23), 30.6 (C-2), 30.0 (C-19), 28.3 (C-16), 26.6 (C-11), 26.3 (C-10), 26.2 (C-7), 25.6 (C-29), 21.3 (C-6), 20.2 (C-9), 19.5 (C-28), 18.5 (C-21), 18.2 (C-18), 17.8 (C-27), 14.2 (C-30).19
(7S,8R,7′S,8′S)-3-Methoxy-3′,4′-methylenedioxy-7,9′- epoxylignane-4,7′,9-triol (12) – Colorless solid; 1H-NMR (400 MHz, acetone-d6): δ 7.42 (s, 4-OH), 6.90 (1H, s, H-2), 6.84 (1H, s, H-2′), 6.75 (1H, d, J = 8.2 Hz, H-6), 6.70 (3H, m, H-5, H-5′, H-6′), 5.91 (2H, d, J = 1.4 Hz, OCH2O), 4.55 (1H, d, J = 7.8 Hz, H-7), 4.52 (1H, d, J = 7.2 Hz, H-7′), 4.15 (1H, dd, J = 8.8, 4.8 Hz, H-9′a), 3.80 (1H, m, H-9′b), 3.78 (3H, s, 3-OCH3), 3.32 (1H, m, H-9a), 3.23 (1H, m, H-9b), 2.49 (1H, m, H-8′), 1.84 (1H, m, H-8); 13CNMR (100 MHz, acetone-d6): δ 148.5 (C-3′), 148.2 (C-3), 146.7 (C-4), 146.6 (C-4′), 139.4 (C-1′), 135.4 (C-1), 121.1 (C-6′), 119.9 (C-6), 115.4 (C-5), 110.7 (C-2), 108.4 (C-5′), 107.9 (C-2′), 101.9 (OCH2O), 84.2 (C-7), 75.7 (C-7′), 70.7 (C-9′), 62.0 (C-9), 56.3 (3-OCH3), 53.4 (C-8), 50.9 (C-8′).20
Luteolin (13) – Yellow amorphous powder; 1H-NMR (400 MHz, CD3OD): δ 7.34 (2H, m, H-2′, H-6′), 6.87 (1H, d, J = 8.8 Hz, H-5′), 6.49 (1H, s, H-3), 6.40 (1H, d, J = 1.4 Hz, H-8), 6.17 (1H, d, J = 1.4 Hz, H-6); 13C-NMR (100 MHz, CD3OD): δ 182.4 (C-4), 164.8 (C-7), 164.5 (C-2), 161.7 (C-5), 157.9 (C-9), 149.5 (C-4′), 145.6 (C-3′), 122.2 (C-1′), 118.8 (C-6′), 115.3 (C-5′), 112.7 (C-2′), 103.9 (C-10), 102.4 (C-3), 98.6 (C-6), 93.5 (C-8).21
(1S,2R)-1-(4-Hydroxy-3-methoxyphenyl)-2-(3-((1E)-3-hydroxy-1-propenyl)-5-methoxyphenoxy)-1,3-propanediol (14) – Colorless solid; 1H-NMR (400 MHz, CD3OD): δ 7.03 (1H, d, J = 1.9 Hz, H-2), 7.01 (1H, s, H-4′), 6.88 (2H, s, H-2′, H-6′), 6.85 (1H, dd, J = 8.1, 1.9 Hz, H-6), 6.74 (1H, d, J = 8.1 Hz, H-5), 6.52 (1H, d, J = 15.9 Hz, H-7′), 6.25 (1H, dt, J = 15.9, 5.8 Hz, H-8′), 4.84 (1H, d, J = 5.7 Hz, H-7), 4.37 (1H, td, J = 5.7, 3.8 Hz, H-8), 4.21 (2H, dd, J = 5.7, 1.3 Hz, H-9′), 3.86 (1H, dd, J = 12.0, 5.7 Hz, H-9a), 3.82 (3H, s, 3′-OCH3), 3.81 (3H, s, 3-OCH3), 3.78 (1H, dd, J = 11.9, 3.8 Hz, H-9b); 13C-NMR (100 MHz, CD3OD): δ 151.9 (C-3′), 148.9 (C-5′), 148.7 (C-3′), 147.0 (C-4′), 134.1 (C-1), 133.0 (C-1′), 131.5 (C-7′), 128.5 (C-8′), 121.0 (C-6), 120.7 (C-2′), 118.9 (C-6′), 115.6 (C-5), 111.9 (C-2), 111.4 (C-4′), 86.2 (C-8), 74.1 (C-7), 63.7 (C-9′), 62.2 (C-9), 56.5 (3′-OCH3), 56.3 (3-OCH3).22
3,6-O-Diferuloyl sucrose (15) – White amorphous powder; 1H-NMR (400 MHz, CD3OD) δ 7.70 (1H, d, J = 15.9 Hz, H-7ʺ), 7.64 (1H, d, J = 15.9 Hz, H-7‴), 7.21 (1H, d, J = 1.4 Hz, H-2ʺ), 7.17 (1H, d, J = 1.5 Hz, H-2‴), 7.12 (1H, dd, J = 8.3, 1.7 Hz, H-6ʺ), 7.07 (1H, dd, J = 8.3, 1.4 Hz, H-6‴), 6.79 (2H, d, J = 8.3 Hz, H-5ʺ, H-5‴), 6.40 (2H, m, H-8ʺ, H-8‴), 5.48 (1H, d, J = 7.9 Hz, H-1′), 5.43 (1H, d, J = 3.7 Hz, H-3), 4.54 (1H, dd, J = 11.9, 7.2 Hz, H-6′a), 4.49 (1H, dd, J = 11.9, 3.8 Hz, H-6′b), 4.43 (1H, t, J = 7.8 Hz, H-2′), 4.15 (1H, m, H-6a), 3.93 (1H, m, H-5), 3.88 (3H, s, 3ʺ-OCH3), 3.87 (3H, s, 3‴-OCH3), 3.84 (1H, m, H-4ʹ), 3.78 (1H, dd, J = 11.7, 4.4 Hz, H-6b), 3.64 (2H, m, H-1a, H-4), 3.58 (1H, d, J = 12.2 Hz, H-1b), 3.41 (1H, dd, J = 9.5, 3.8 Hz, H-3′), 3.37 (1H, t, J = 9.0 Hz, H-5′); 13C-NMR (100 MHz, CD3OD): δ 168.1 (C-9ʺ), 167.3 (C-9‴), 149.7 (C-4ʺ, C-4‴), 148.4 (C-3ʺ), 148.4 (C-3‴), 146.9 (C-7ʺ), 146.3 (C-7‴), 127.7 (C-1ʺ), 127.7 (C-1‴), 123.3 (C-6ʺ), 123.3 (C-6‴), 115.5 (C-5ʺ, C-5‴), 114.2 (C-8ʺ), 114.0 (C-8‴), 111.1 (C-2ʺ), 111.0 (C-2‴), 104.1 (C-2), 92.1 (C-1′), 80.3 (C-5), 78.2 (C-3), 74.0 (C-4), 74.0 (C-2′), 73.4 (C-3′), 72.2 (C-4′), 70.5 (C-5′), 65.7 (C-1), 64.2 (C-6′), 61.6 (C-6), 55.6 (3ʺ-OCH3), 55.5 (3‴-OCH3).23
3,6-O-Diferuloyl-6ʹ-O-acetyl sucrose (16)‒White amorphous powder; 1H-NMR (400 MHz, CD3OD): δ 7.68 (1H, d, J = 15.9 Hz, H-7ʺ), 7.61 (1H, d, J = 15.9 Hz, H-7‴), 7.21 (1H, s, H-2ʺ), 7.14 (1H, s, H-2‴), 7.09 (1H, d, J = 8.2 Hz, H-6ʺ), 7.05 (1H, d, J = 8.1 Hz, H-6‴), 6.78 (2H, m, H-5ʺ, 5‴), 6.41 (1H, dd, J = 15.9, 1.3 Hz, H-8ʺ), 6.36 (1H, dd, J = 15.9, 1.3 Hz, H-8‴), 5.48 (2H, m, H-3, H-1′), 4.51 (4H, m, H-6a, H-2′, H-6′a), 4.16 (1H, m, H-4′), 4.09 (2H, m, H-5, H-6b), 3.86 (3H, s, 3ʺ-OCH3), 3.86 (3H, s, 3‴-OCH3), 3.60 (3H, m, H-1, H-4), 3.42 (1H, dd, J = 9.7, 3.8 Hz, H-3′), 3.25 (1H, t, J = 9.4 Hz, H-5′), 2.07 (3H, s, COCH3); 13C-NMR (100 MHz, CD3OD) δ (ppm): 172.7 (COCH3), 168.1 (C-9ʺ), 168.2 (C-9‴), 150.7 (C-4ʺ), 150.6 (C-4‴), 149.3 (C-3ʺ), 149.3 (C-3‴), 147.9 (C-7ʺ), 147.1 (C-7‴), 127.7 (C-1ʺ), 127.6 (C-1‴), 124.3 (C-6ʺ), 124.2 (C-6‴), 116.5 (C-5ʺ), 116.4 (C-5‴), 115.2 (C-8ʺ), 114.8 (C-8‴), 111.9 (C-2ʺ), 111.6 (C-2‴), 104.8 (C-2), 92.5 (C-1′), 81.2 (C-5), 78.9 (C-3), 74.9 (C-4), 74.3 (C-2′), 73.0 (C-3′), 72.1 (C-4′), 71.9 (C-5′), 65.7 (C-1), 65.6 (C-6′), 65.5 (C-6), 56.5 (3ʺ-OCH3), 56.5 (3‴-OCH3), 20.7 (COCH3).23
Luteolin 4′-O-glucoside (17) – Yellow solid; 1H-NMR (500 MHz, DMSO-d6): δ 12.90 (1H, s, 5-OH), 7.52 (1H, dd, J = 8.2, 2.3 Hz, H-6′), 7.49 (1H, d, J = 2.2 Hz, H-2′), 7.24 (1H, d, J = 8.6 Hz, H-5′), 6.82 (1H, s, H-3), 6.50 (1H, d, J = 2.1 Hz, H-8), 6.19 (1H, d, J = 2.1 Hz, H-6), 4.88 (1H, d, J = 7.3 Hz, H-1ʺ), 3.73 (1H, d, J = 10.0 Hz, H-6ʺb), 3.48 (1H, dd, J = 11.4, 5.3 Hz, H-6ʺa), 3.40‒3.16 (4H, br, H-2ʺ‒H-5ʺ); 13C-NMR (125 MHz, DMSO-d6): δ 182.3 (C-4), 164.8 (C-7), 163.7 (C-2), 162.0 (C-5), 157.9 (C-9), 149.1 (C-4′), 147.4 (C-3′), 125.2 (C-1′), 119.0 (C-6′), 116.5 (C-5′), 114.1 (C-2′), 104.5 (C-10), 104.3 (C-3), 101.7 (C-1ʺ), 99.4 (C-6), 94.5 (C-8), 77.8 (C-5ʺ), 76.3 (C-3ʺ), 73.8 (C-2ʺ), 70.3 (C-4ʺ), 61.2 (C-6ʺ).24
7R,8R-Dihydrodehydrodiconiferyl alcohol-9-O-β-Dglucopyranoside (18) – Colorless solid; 1H--NMR (500 MHz, CD3OD): δ 7.02 (1H, d, J = 1.9 Hz, H-2), 6.87 (1H, dd, J = 8.2, 1.9 Hz, H-6), 6.82 (1H, s, H-2′), 6.78 (1H, d, J = 8.2 Hz, H-5), 6.74 (1H, s, H-6′), 5.60 (1H, d, J = 6.4 Hz, H-7), 4.38 (1H, d, J = 7.8 Hz, H-1ʺ), 4.13 (1H, dd, J = 9.6, 8.0 Hz, Ha-9), 3.90 (1H, m, H-9b), 3.86 (1H, m, Ha-6ʺ), 3.70 (1H, dd, J = 11.8, 5.3 Hz, H-6ʺb), 3.66 (1H, m, H-8), 3.87 (3H, s, 3′-OCH3), 3.85 (3H, s, 3-OCH3), 3.54 (2H, m, H-9′), 3.31‒3.23 (4H, m, H-2ʺ‒H-5ʺ), 2.64 (2H, m, CH2-7′), 1.83 (2H, m, CH2-8′); 13C-NMR (125 MHz, CD3OD): δ 149.0 (C-3), 147.4 (C-4′), 147.4 (C-4), 145.2 (C-3′), 137.0 (C-1′), 134.6 (C-1), 129.7 (C-5′), 119.8 (C-6), 118.2 (C-2′), 116.1 (C-6′), 114.1 (C-5), 110.8 (C-2), 104.2 (C-1ʺ), 89.2 (C-7), 78.2 (C-3ʺ), 78.0 (C-5ʺ), 75.1 (C-2ʺ), 72.3 (C-4ʺ), 71.6 (C-9), 62.7 (C-9′), 62.2 (C-6ʺ), 56.7 (3′-OCH3), 56.4 (3-OCH3), 52.9 (C-8), 35.8 (C-7′), 32.7 (C-8′).25
3-O-Feruloyl-6ʹ-O-acetyl sucrose (19) – Pale yellow solid; 1H-NMR (400 MHz, CD3OD): δ 7.68 (1H, d, J = 15.9 Hz, H-7ʺ), 7.23 (1H, s, H-2ʺ), 7.11 (1H, d, J = 8.2 Hz, H-6ʺ), 6.79 (1H, d, J = 8.2 Hz, H-5ʺ), 6.41 (1H, dd, J = 15.9, 1.3 Hz, H-8ʺ), 5.44 (2H, m, H-3, H-1′), 4.49 (1H, d, J = 10.1 Hz, H-6′a), 4.33 (1H, t, J = 8.0 Hz, H-2′), 4.12 (2H, m, H-4′, H-6′b), 3.93 (1H, m, H-5), 3.88 (3H, s, 3ʺ- OCH3), 3.85 (1H, m, H-6a), 3.79 (2H, m, H-6b), 3.59 (3H, m, H-1, H-4), 3.41 (1H, dd, J = 9.7, 3.6 Hz, H-3′), 3.25 (1H, m, H-5′), 2.07 (3H, s, COCH3); 13C-NMR (100 MHz, CD3OD): δ 172.7 (COCH3), 168.1 (C-9ʺ), 150.5 (C-4ʺ), 149.2 (C-3ʺ), 147.5 (C-7ʺ), 127.5 (C-1ʺ), 124.0 (C-6ʺ), 116.2 (C-5ʺ), 114.8 (C-8ʺ), 111.9 (C-2ʺ), 104.6 (C-2), 92.7 (C-1′), 84.1 (C-5), 79.3 (C-3), 74.7 (C-4), 73.9 (C-2′), 72.8 (C-3′), 72.0 (C-4′), 71.5 (C-5′), 65.3 (C-1), 65.1 (C-6′), 63.5 (C-6), 56.3 (3ʺ-OCH3), 20.7 (COCH3).26
7R,8R-Dihydro-9′-hydroxyl-3′-methoxyl-8-hydroxymethyl-7-(4-hydroxy-3-methoxyphenyl)-1′-benzofuranpropanol 9′-O-β-D-glucopyranoside (20) – Colorless solid; 1HNMR (500 MHz, CD3OD): δ 6.95 (1H, s, H-2), 6.82 (1H, dd, J = 8.2, 1.7 Hz, H-6), 6.77 (1H, s, H-2′), 6.78 (1H, d, J = 8.2 Hz, H-5), 6.77 (1H, s, H-6′), 5.51 (1H, d, J = 6.1 Hz, H-7), 4.27 (1H, d, J = 7.8 Hz, H-1ʺ), 4.27 (1H, d, J = 9.6, 8.0 Hz, H-9a), 3.90 (1H, m, H-9b), 3.86 (1H, m, H-6ʺa), 3.70 (1H, dd, J = 11.8, 5.3 Hz, H-6ʺb), 3.66 (1H, m, H-8), 3.87 (3H, s, 3′-OCH3), 3.84 (3H, s, 3-OCH3), 3.60 (2H, t, J = 6.5 Hz, H-9′), 3.31‒3.20 (4H, m, H-2ʺ‒H-5ʺ), 2.70 (2H, t, J = 7.5 Hz, H-7′), 1.93 (2H, m, H-8′); 13CNMR (125 MHz, CD3OD): δ 149.1 (C-3), 147.5 (C-4′), 147.5 (C-4), 145.2 (C-3′), 136.8 (C-1′), 134.8 (C-1), 129.8 (C-5′), 119.7 (C-6), 118.0 (C-2′), 116.1 (C-6′), 114.2 (C-5), 110.6 (C-2), 104.5 (C-1ʺ), 88.9 (C-7), 78.2 (C-3ʺ), 77.9 (C-5ʺ), 75.1 (C-2ʺ), 71.7 (C-4ʺ), 69.9 (C-9′), 65.0 (C-9), 62.8 (C-6ʺ), 56.8 (3′-OCH3), 56.4 (3-OCH3), 55.4 (C-8), 32.9 (C-7′), 32.9 (C-8′).27
Isorhamnetin-3-O-glucoside (21) – Yellow solid; 1HNMR (400 MHz, CD3OD): δ 7.88 (1H, s, H-2′), 7.53 (1H, d, J = 8.2 Hz, H-6′), 6.86 (1H, d, J = 8.2 Hz, H-5′), 6.30 (1H, s, H-8), 6.13 (1H, s, H-6), 5.36 (1H, d, J = 7.3 Hz, H-1ʺ), 3.91 (3H, s, 3′-OCH3), 3.73 (1H, d, J = 10.1 Hz, H-6ʺb), 3.56 (1H, dd, J = 11.9, 5.3 Hz, H-6ʺa), 3.31‒3.25 (4H, br, H-2ʺ‒H-5ʺ); 13C-NMR (100 MHz, CD3OD): δ 179.2 (C-4), 165.7 (C-7), 162.8 (C-5), 158.4 (C-9), 158.2 (C-2), 150.7 (C-4′), 148.2 (C-3′), 135.2 (C-3), 123.7 (C-1′), 122.9 (C-6′), 115.8 (C-5′), 114.2 (C-2′), 105.6 (C-10), 103.7 (C-1ʺ), 99.8 (C-6), 94.7 (C-8), 78.3 (C-5ʺ), 77.9 (C-3ʺ), 75.8 (C-2ʺ), 71.4 (C-4ʺ), 62.5 (C-6ʺ), 56.6 (3′-OCH3).28
Isorhamnetin-3-O-rutinoside (22) – Yellow solid; 1HNMR (400 MHz, CD3OD): δ 7.91 (1H, d, J = 1.7 Hz, H-2′), 7.59 (1H, d, J = 8.5, 1.7 Hz, H-6′), 6.88 (1H, d, J = 8.5 Hz, H-5′), 6.37 (1H, d, J = 1.6 Hz, H-8), 6.18 (1H, d, J = 1 .6 Hz, H-6), 5 .21 (1H, d , J = 7.2 Hz, H-1ʺ), 4.50 (1H, s, H-1‴), 3.92 (3H, s, 3′-OCH3), 3 .79 (1H, d , J = 10.9 Hz, H-6ʺb), 3.46‒3.23 (9H, H-2ʺ‒H-6ʺa, H-2‴‒H-5‴), 1.07 (3H, d, J = 6.2 Hz, H-6‴); 13C-NMR (100 MHz, CD3OD): δ 179.2 (C-4), 165.9 (C-7), 162.9 (C-5), 158.7 (C-9), 158.7 (C-2), 150.7 (C-4′), 148.2 (C-3′), 135.3 (C-3), 123.8 (C-1′), 122.9 (C-6′), 116.0 (C-5′), 114.4 (C-2′), 105.6 (C-10), 104.3 (C-1‴), 102.4 (C-1ʺ), 99.8 (C-6), 94.8 (C-8), 78.0 (C-5ʺ), 77.3 (C-3ʺ), 75.8 (C-2ʺ), 73.7 (C-5‴), 72.3 (C-2‴), 72.0 (C-4‴), 71.5 (C-4ʺ), 69.7 (C-3‴), 68.4 (C-6ʺ), 56.6 (3′-OCH3), 17.8 (C-6‴).29
3,5-O-Dicaffeoylquinic acid (23) – Pale yellow solid; 1H-NMR (400 MHz, CD3OD): δ 7.49 (1H, d, J = 15.8 Hz, H-7′), 7.47 (1H, d, J = 15.8 Hz, H-7ʺ), 6.97 (1H, s, H-2′), 6.94 (1H, s, H-2ʺ), 6.84 (2H, m, H-6′, H-6ʺ), 6.67 (2H, d, J = 8.1 Hz, H-5′, H-5ʺ), 6.30 (1H, d, J = 15.8 Hz, H-8′), 6.18 (1H, d, J = 15.8 Hz, H-8ʺ), 5.41 (1H, m, H-3), 5.30 (1H, m, H-5), 3.82 (1H, dd, J = 9.6, 3.3 Hz, H-4), 2.18 (1H, dd, J = 14.7, 2.4 Hz, H-6a), 2.02 (3H, m, H-2, H-6b); 13C-NMR (100 MHz, CD3OD): δ 181.2 (C-7), 169.3 (C-9′), 168.9 (C-9ʺ), 149.4 (C-4′), 149.3 (C-4ʺ), 147.0 (C-3′), 146.9 (C-3ʺ), 146.7 (C-7′), 146.7 (C-7ʺ), 128.0 (C-1′), 127.9 (C-1ʺ), 123.0 (C-6′), 123.0 (C-6ʺ), 116.5 (C-5′), 116.5 (C-5ʺ), 115.9 (C-8ʺ), 115.4 (C-8‴), 115.2 (C-2′, 2ʺ), 76.5 (C-1), 74.2 (C-5), 72.9 (C-4), 72.3 (C-3), 40.4 (C-2), 37.4 (C-6).30
Cell culture – RAW 264.7 cells (ATCC, Rockville, MD, USA) were cultured in DMEM medium containing 10% FBS and penicillin (100 units/mL) at 37oC in a 5% CO2-humidified air environment. HeLa and HL-60 cancer cell lines (ATCC, Rockville, MD, USA) were cultured in RPMI medium containing 10% FBS and penicillin (100 units/mL) at 37oC in a 5% CO2-humidified air environment.
Cell viability and griess assay – RAW 264.7 cells were seeded in 96-well plates at a density of 1 × 105 cells in each well. After 3 hours, the cells were pre-treated with varying concentrations of the test samples for 30 minutes, followed by incubation for 24 hours with or without 1 μg/mL of LPS. The concentration of NO in the culture supernatant was then measured using the Griess reaction.31,32 Cell viabilities (for RAW 264.7 macrophages, HeLa, and HL-60 cancer cells) were assessed using the MTT assay.32,33 Significant differences were determined by comparing the results to the untreated control group.
Statistical analysis – All experimental bioassay data were performed as the mean ± standard error (SE) of the mean from at least three independent experiments. Oneway analysis of variance (ANOVA) and Dunnett’s test were applied to evaluate statistical significance. p values of less than 0.05 were considered to be statistically significant.
Results and Discussion
Twenty-three known compounds including taraxasteryl acetate (1),11 taraxast-20(30)-en-3-one (2),12 neoilexonol acetate (3),13 epifriedelanol (4),14 stigmast 7-en-3-one (5),15 sesamin (6),16 7-keto-β-sitosterol (7),17 ergosterol peroxide (8),18 9,19-cyclolanost-23-ene-3β,24-diol (9),19 (24α/β)-9,19-cyclolanost-25-ene-3β,24-diol (10/11),19 (7S,8R,7′S,8′S)-3-methoxy-3′,4′-methylenedioxy-7,9′-epoxylignane-4,7′,9-triol (12),20 luteolin (13),21 (1S,2R)-1-(4-hydroxy-3-methoxyphenyl)-2-(3-((1E)-3-hydroxy-1-propenyl)-5-methoxyphenoxy)-1,3-propanediol (14),22 3,6-O-diferuloyl sucrose (15),23 3,6-O-diferuloyl-6ʹ-O-acetyl sucrose (16),23 luteolin 4′-O-glucoside (17),24 7R,8R-dihydrodehydrodiconiferyl alcohol-9-O-β-D-glucopyranoside (18),25 3-O-feruloyl-6ʹ-O-acetyl sucrose (19),26 7R,8R-dihydro-9′-hydroxyl-3′- methoxyl-8-hydroxymethyl-7-(4-hydroxy-3-methoxyphenyl)-1′-benzofuranpropanol 9′-O-β-D-glucopyranoside (20),27 isorhamnetin-3-O-glucoside (21),28 isorhamnetin-3-O-rutinoside (22),29 and 3,5-O-dicaffeoylquinic acid (23)30 were isolated from aerial parts of A. alpina L. (Fig. 1). Their structures were determined by comparing their 1D NMR spectral data with those in the literature. Among them, compounds 2, 3, 5, 7, 12, 14, 18 and 20 were reported from A. alpina for the first time.
Compound 8 was obtained as a colorless oil. Its 1H NMR spectrum displayed signals in the high-field region, indicating the presence of six methyl groups typical of a steroid skeleton, including four doublet methyls at δH 0.98 (3H, d, J = 6.6 Hz, H-21), 0.89 (3H, J = 6.8 Hz, CH3-28), and 0.79 (6H, d, J = 3.2 Hz, CH3-26, CH3-27) and two singlet ones at δH 0.86 (3H, s, CH3-19) and 0.80 (3H, s, H-18). An oxygenated methine proton δH 3.93 (1H, m), a characteristic of 3-OH group in steroids, was also detected. Additionally, four olefinic protons characteristic of one cis-double bond at δH 6.48 (1H, d, J = 8.5 Hz, H-7) and 6.22 (1H, d, J = 8.5 Hz, H-6) and one trans-double bond at δH 5.20 (1H, dd, J = 15.2, 7.4 Hz, H-22) and 5.12 (1H, dd, J = 15.2, 8.2 Hz, H-23) were observed. The 13C NMR spectrum of 8 showed 28 carbon signals (Fig. S8), with four olefinic carbons belonging to one cis- and one trans-double bond at δC 135.3 (C-6), 135.1 (C-22), 132.1 (C-23), and 130.6 (C-7), suggesting that 8 is an ergosterol derivative.34 Especially, two oxygenated sp3 quaternary carbons at δC 82.1 (C-5) and 79.3 (C-8) indicated the presence of a peroxide group, deducing 8 as ergosterol peroxide. A comparison of the NMR data of 8 with those in the previous report confirmed that 8 was ergosterol peroxide.18
All the isolated compounds were tested for their ability to inhibit NO production in LPS-stimulated RAW 264.7 mouse macrophages. Following LPS treatment, NO levels in the culture medium increased significantly. Our results demonstrated that luteolin (13), a well-known flavone from the genus Achillea, showed the most potent inhibitory effect with an IC50 value of 8.69 ± 0.12 μM (Fig. 2). This was followed by sterols, ergosterol peroxide (8) (Fig. 2) and 7-keto-β-sitosterol (7), which had IC50 values of 18.39 ± 0.27 μM and 28.37 ± 0.75 μM, respectively. Interestingly, these compounds exhibited significantly stronger inhibition than the positive control, quercetin (IC50 = 33.21 μM ± 0.55).35 Additionally, stigmast-7-en-3-one (5) showed moderate inhibition of NO production with an IC50 value of 43.17 ± 0.27 μM, while cycloartane isomers 10 a nd 11 exhibited weak inhibitory activity with IC50 values around 90 μM. The other compounds were inactive at a concentration of 100 μM (Table 1).
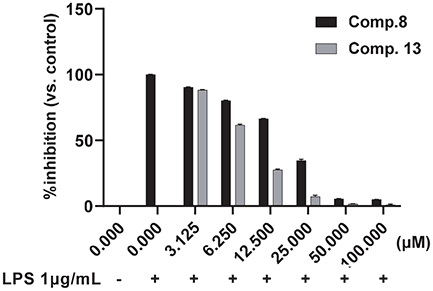
Effect of compounds 8 and 13 on the LPS-induced NO production in RAW 264.7 cells. RAW 264.7 cells were pretreated with 1‒100 μM of 8 and 13 for 30 min, then incubated with 1 μg/mL LPS for an additional 24 h. DMSO was used as a vehicle. NO was measured by Griess assay. The data are expressed as the mean ± SE (n = 3).
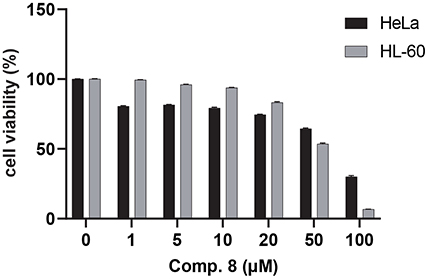
Cytotoxic effect of compound 8 on HeLa and HL-60 cancer cell lines. HeLa and HL-60 cells were treated with 1‒100 μM of 8 and incubated for 48 h. DMSO was used as a vehicle. Cell viability was measured by MTT assay. The data are expressed as the mean ± SE (n = 3).
The MTT assay was used to evaluate the cytotoxicity of the isolated compounds on RAW 264.7 cells at concentrations ranging from 1–100 μM. The cell viability was largely unaffected in the presence of compounds 5, 7, 8, 10, 11, and 13, deducing these compounds, at noncytotoxic levels (≤ 100 μM), effectively prevented the LPS-induced inflammatory response in RAW 264.7 cells, as indicated by NO production.
The cytotoxicity of all isolated compounds on HeLa and HL-60 cells was assessed using the MTT assay (Table 2). Among them, ergosterol peroxide (8) exhibited significant cytotoxicity on both HeLa and HL-60 cells, with EC50 values of 72.85 and 53.57 μM, respectively. In contrast, compounds 5 and 10 selectively inhibited the proliferation of HL-60 cells, showing EC50 values of 78.33 and 58.83 μM, respectively. Compound 11 was only cytotoxic to HeLa cells, with an EC50 value of 68.45 μM.
The overall bioactivity assessment results indicated that attaching sugar units to any position on the isolated flavonoids significantly reduced their NO production inhibitory activity, evidenced by the differing inhibitory effects between compound 13 and compounds 17, 21, and 22. Both cycloartane isomers 10/11 exhibited similar inhibitory activity against excessive NO production; however, the 24α-isomer (10) induced cell death in HL-60 cells, whereas the other isomer (11-24β) only caused cell death in HeLa cells. This suggested that configuration changes could also alter their biological effects. While the isolated pentacyclic triterpenes (1‒4) showed almost no activity at 100 μM, the sterols (5, 7, and 8) exhibited notable activity. In particular, our experiments demonstrated both the anti-inflammatory and anti-cancer effects of ergosterol peroxide (8), a steroid characterized by a unique peroxide group - a key responsible for its pharmacological effects. This compound, previously found in various fungi and medicinal herbs, has been shown to offer numerous health benefits such as anti-oxidant,36 anti-inflammatory,37 anti-cancer,38 anti-viral,39 anti-obesity,40 and anti-fungal41 activities. Our results further supported its diverse pharmacological activities.
In summary, a total of 23 secondary metabolites, including 4 pentacyclic triterpenes (1‒4), 3 steroids (5, 7, and 8), 3 cycloartanes (9‒11), 5 lignans (6, 12, 14, 20, and 18), 4 flavonoids (13, 17, 21, and 22), 3 feruloyl sucroses (16, 18, and 19), and 1 caffeoyl quinic acid (23), were isolated and structurally identified from the aerial parts of A. alpina. This report marked the first isolation of compounds 2, 3, 5, 7, 12, 14, 18 and 20 from this plant. The bioactivity studies revealed that ergosterol peroxide (8) and luteolin (13) are potent inhibitors of NO production in LPSstimulated RAW 264.7 cells. Additionally, ergosterol peroxide (8) was also shown to significantly induce cell death in the HeLa and HL-60 cancer cell lines. These findings provided insights into the phytochemical characteristics of A. alpina, and the anti-inflammatory and anticancer activities of its metabolites.
Acknowledgments
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT (NRF-2021R1A2C2011940), Korea.
Conflicts of Interest
The authors declare no competing financial interest.
References
-
Barda, C.; Grafakou, M.-E.; Tomou, E.-M.; Skaltsa, H. Sci. Pharm. 2021, 89, 50.
[https://doi.org/10.3390/scipharm89040050]

-
Saeidnia, S.; Gohari, A.; Mokhber-Dezfuli, N.; Kiuchi, F. Daru. 2011, 19, 173–186.
[https://doi.org/10.1016/B978-0-444-59514-0.00006-7]

-
Zhang, Q.; Zhou, Q.; Huo, C.; Wang, Y.; Wu, Y.; Zhang, M.; Li, L.; Jin, S.; Shi, Q.; Gu, Y. Chem. Nat. Compd. 2019, 55, 337–339.
[https://doi.org/10.1007/s10600-019-02683-x]

-
Lee, H. J.; Sim, M. O.; Woo, K. W.; Jeong, D.-E.; Jung, H. K.; An, B.; Cho, H. W. Chem. Biodivers. 2019, 16, e1900033.
[https://doi.org/10.1002/cbdv.201900033]

-
Xue, G.-M.; Zhao, C.-G.; Xue, J.-F.; Du, K.; Duan, J.-J.; Pan, H.; Li, M.; Chen, H.; Sun, Y.-J.; Feng, W.-S.; Ma, T.; Zhang, W.-D. Phytochemistry 2022, 202, 113297.
[https://doi.org/10.1016/j.phytochem.2022.113297]

-
Xue, G.-M.; Zhao, C.-G.; Xue, J.-F.; Duan, J.-J.; Pan, H.; Jia, Y.-Y.; Du, K.; Zhi, Y.-L.; Feng, W.-S. Fitoterapia 2023, 166, 105472.
[https://doi.org/10.1016/j.fitote.2023.105472]

-
Le, T. T.; Ha, M. T.; Lee, G. S.; Nguyen, V. P.; Kim, C. S.; Kim, J. A. ; Min, B. S. Phytochemistry 2025, 229, 114269.
[https://doi.org/10.1016/j.phytochem.2024.114269]

-
Li, X.; Wu, X.; Guo, Q.; Liu, C.; Yue, H.; Tian, J.; Liu, Z.; Xiao, M.; Li, A.; Ning, Z.; Zan, K. Biochem. Syst. Ecol. 2022, 101, 104381.
[https://doi.org/10.1016/j.bse.2022.104381]

-
Le, T. T.; Tran, T. T.; Ha, M. T.; Kim, J. A.; Min, B. S. Biochem. Syst. Ecol. 2025, 119, 104924.
[https://doi.org/10.1016/j.bse.2024.104924]

-
Li, X.-W.; Yue, H.-C.; Wu, X.; Guo, Q.; Tian, J.-Y.; Liu, Z.-Y.; Xiao, M.; Li, X.-X.; Yu, L.; Li, A.; Ning, Z.-Q.; Zan, K.; Chen, X.-Q. Chem. Biodivers. 2022, 19, e202200218.
[https://doi.org/10.1002/cbdv.202200218]

-
Khalilov, L. M.; Khalilova, A. Z.; Shakurova, E. R.; Nuriev, I. F.; Kachala, V. V.; Shashkov, A. S.; Dzhemilev, U. M. Chem. Nat. Compd. 2003, 39, 285–288.
[https://doi.org/10.1023/A:1025478720459]

-
Akhmetova, V. R.; Shakurova, E. R.; Khalilov, L. M. Russ. J. Org. Chem. 2009, 45, 621–623.
[https://doi.org/10.1134/S1070428009040265]

-
Ogawa, S.; Wakatsuki, Y.; Makino, M.; Fujimoto, Y.; Yasukawa, K.; Kikuchi, T.; Ukiya, M.; Akihisa, T.; Iida, T. Chem. Phys. Lipids 2010, 163, 165–171.
[https://doi.org/10.1016/j.chemphyslip.2009.10.012]

-
Kundu, J. K.; Rouf, A. S.; Hossain, N. M.; Hasan, C. M.; Rashid, M. A. Fitoterapia 2000, 71, 577–579.
[https://doi.org/10.1016/S0367-326X(00)00191-X]

-
Wu, F.-E.; Koike, K.; Nikaido, T.; ISHII, K.; Ohmoto, T.; Ikeda, K. Chem. Pharm. Bull. 1990, 38, 2281–2282.
[https://doi.org/10.1248/cpb.38.2281]

-
Dar, A. A.; Verma, N. K.; Arumugam, N. Ind. Crops Prod. 2015, 64, 201–208.
[https://doi.org/10.1016/j.indcrop.2014.10.026]

-
Pettit, G. R.; Numata, A.; Cragg, G. M.; Herald, D. L.; Takada, T.; Iwamoto, C.; Riesen, R.; Schmidt, J. M.; Doubek, D. L.; Goswami, A. J. Nat. Prod. 2000, 63, 72–78.
[https://doi.org/10.1021/np990346r]

-
El-Sherif, N. F.; Ahmed, S. A.; Ibrahim, A. K.; Habib, E. S.; El-Fallal, A. A.; El-Sayed, A. K. A.; Wahba, A. E. Int. J. Med. Mushrooms 2020, 22, 389–396.
[https://doi.org/10.1615/IntJMedMushrooms.2020034223]

-
Paula, V. F.; Barbosa, L. C. A.; Errington, W.; Howarth, O. W.; Cruz, M. P. J. Braz. Chem. Soc. 2002, 13, 276–280.
[https://doi.org/10.1590/S0103-50532002000200022]

-
Lee, J.; Lee, Y. J.; Oh, S.-M.; Yi, J.-M.; Kim, N. S.; Bang, O.-S. Molecules 2013, 19, 122–138.
[https://doi.org/10.3390/molecules19010122]

-
Lin, L.-C.; Pai, Y.-F.; Tsai, T.-H. J. Agric. Food Chem. 2015, 63, 7700–7706.
[https://doi.org/10.1021/jf505848z]

-
Greca, M. D.; Molinaro, A.; Monaco, P.; Previtera, L. Phytochemistry 1994, 35, 777–779.
[https://doi.org/10.1016/S0031-9422(00)90604-6]

-
Sun, X.; Zimmermann, M. L.; Campagne, J. M.; Sneden, A. T. J. Nat. Prod. 2000, 63, 1094–1097.
[https://doi.org/10.1021/np000055e]

-
Lin, Y.; Liu, P.-G.; Liang, W.-Q.; Hu, Y.-J.; Xu, P.; Zhou, J.; Pu, J.-B.; Zhang, H.-J. Phytomedicine 2018, 41, 54–61.
[https://doi.org/10.1016/j.phymed.2018.02.002]

-
Kumar, P.; Dev, K.; Sharma, K.; Sahai, M.; Maurya, R. Nat. Prod. Res. 2019, 33, 233–238.
[https://doi.org/10.1080/14786419.2018.1443099]

-
Cho, J.-G.; Cha, B.-J.; Seo, W.-D.; Jeong, R.-H.; Shrestha, S.; Kim, J.-Y.; Kang, H.-C.; Baek, N.-I. Chem. Nat. Compd. 2015, 51, 1094–1098.
[https://doi.org/10.1007/s10600-015-1500-8]

-
Dong, L.-P.; Ni, W.; Dong, J.-Y.; Li, J.-Z.; Chen, C.-X.; Liu, H.-Y. Molecules 2006, 11, 1009–1014.
[https://doi.org/10.3390/11121009]

-
Wang, D.-M.; Pu, W.-J.; Wang, Y.-H.; Zhang, Y.-J.; Wang, S.-S. Molecules 2012, 17, 4595–4603.
[https://doi.org/10.3390/molecules17044595]

-
Boubaker, J.; Bhouri, W.; Ben Sghaier, M.; Ghedira, K.; Dijoux Franca, M. G.; Chekir-Ghedira, L. Cell Prolif. 2011, 44, 453–461.
[https://doi.org/10.1111/j.1365-2184.2011.00772.x]

-
Garayev, E.; Di Giorgio, C.; Herbette, G.; Mabrouki, F.; Chiffolleau, P.; Roux, D.; Sallanon, H.; Ollivier, E.; Elias, R.; Baghdikian, B. J. Ethnopharmacol. 2018, 226, 176–184.
[https://doi.org/10.1016/j.jep.2018.08.005]

-
Lee, S.; Choi, S. Y.; Choo, Y.-Y.; Kim, O.; Tran, P. T.; Dao, C. T.; Min, B.-S.; Lee, J.-H. Int. Immunopharmacol. 2015, 28, 328–336.
[https://doi.org/10.1016/j.intimp.2015.06.015]

- Le, T. T.; Ha, M. T.; Han, K.-H.; Kim, Y.-B.; Kim, J. A.; Min, B. S. Chem. Biodiver. 2022, 19, e202100986.
-
Phong, N. V.; Chae, H. Y.; Oanh, V. T.; Min, B. S.; Kwon, M. J.; Kim, J. A. Nat. Prod. Sci. 2023, 29, 171–181.
[https://doi.org/10.20307/nps.2023.29.3.171]

-
Nowak, R.; Drozd, M.; Mendyk, E.; Lemieszek, M.; Krakowiak, O.; Kisiel, W.; Rzeski, W.; Szewczyk, K. Molecules 2016, 21, 946.
[https://doi.org/10.3390/molecules21070946]

-
Vu, N. K.; Le, T. T.; Woo, M. H.; Min, B. S. Nat. Prod. Sci. 2021, 27, 176–182.
[https://doi.org/10.20307/nps.2021.27.3.176]

-
Dupont, S.; Fleurat-Lessard, P.; Cruz, R. G.; Lafarge, C.; Grangeteau, C.; Yahou, F.; Gerbeau-Pissot, P.; Abrahão Júnior, O.; Gervais, P.; Simon-Plas, F.; Cayot, P.; Beney, L. Antioxidants 2021, 10, 1024.
[https://doi.org/10.3390/antiox10071024]

-
Kobori, M.; Yoshida, M.; Ohnishi-Kameyama, M.; Shinmoto, H. Br. J. Pharmacol. 2007, 150, 209–219.
[https://doi.org/10.1038/sj.bjp.0706972]

-
Wu, H.-Y.; Yang, F.-L.; Li, L.-H.; Rao, Y. K.; Ju, T.-C.; Wong, W.- T.; Hsieh, C.-Y.; Pivkin, M. V.; Hua, K.-F.; Wu, S.-H. Sci. Rep. 2018, 8, 17956.
[https://doi.org/10.1038/s41598-018-36411-2]

-
Zhou, B.; Liang, X.; Feng, Q.; Li, J.; Pan, X.; Xie, P.; Jiang, Z.; Yang, Z. Eur. J. Pharmacol. 2019, 860, 172543.
[https://doi.org/10.1016/j.ejphar.2019.172543]

-
Jeong, Y.-U.; Park, Y.-J. Int. J. Mol. Sci. 2020, 21, 460.
[https://doi.org/10.3390/ijms21020460]

-
Daroodi, Z.; Taheri, P.; Tarighi, S.; Iranshahi, M.; Akaberi, M. J. Appl. Microbiol. 2024, 135, lxae031.
[https://doi.org/10.1093/jambio/lxae031]